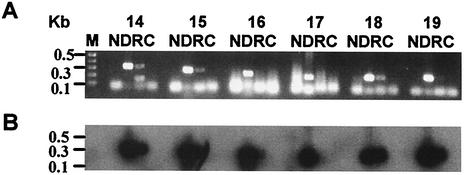
FIG. 7.
RT-PCR analysis of E. chaffeensis isolate Arkansas RNA extracted from in vitro-cultured organisms. Purified RNA was digested with RQ1 DNase for 1 h to remove any residual genomic DNA. DNA-free RNA was used for the RT-PCR assay by using gene-specific primer sets for genes 14 to 19. The primers were designed from variable regions and are listed in Table 2. Lane N contained all reagents for the RT-PCR assay, except RNA. Lane D is the same as lane N plus genomic DNA. Lane R is same as lane N but with DNase-treated RNA as the template. Lane C is same as lane R except that the reverse transcriptase is omitted. (A) Amplified products resolved on a 1.2% agarose gel and detected after being stained with ethidium bromide. (B) RT-PCR products transferred to a nylon membrane and hybridized with a 32P-labeled 28-kDa gene probe. St. Vincent, Wakulla, and Liberty isolates were similarly analyzed.