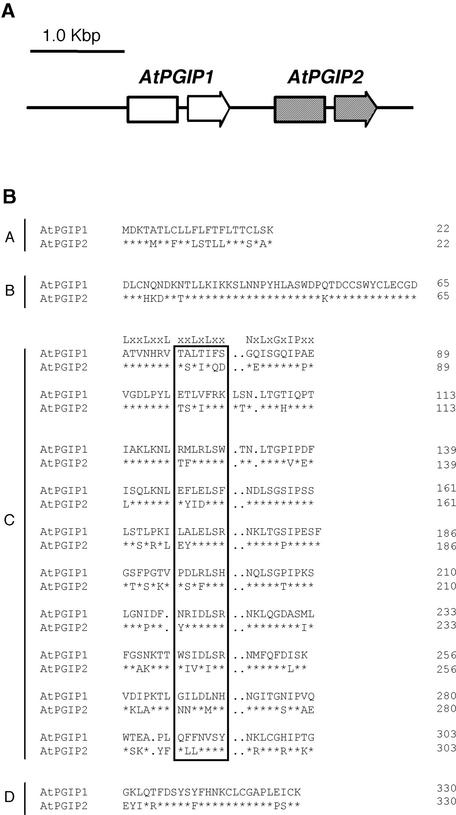
Figure 1.
Genomic Organization of AtPGIP1 and AtPGIP2 and Comparison of the Predicted Amino Acid Sequences.
(A) Genomic organization of AtPGIP1 and AtPGIP2. Arrows represent AtPGIP1 and AtPGIP2 open reading frames, each interrupted by an intron.
(B) Sequence alignment of AtPGIP1 and AtPGIP2. The predicted amino acid sequences were aligned using the CLUSTAL W method (Higgins and Sharp, 1988). Typical PGIP domains are indicated (A, signal peptide; B, presumed N terminus of the mature protein; C, 10 LRR modules; D, C terminus). The consensus sequence for extracytoplasmic LRR is indicated above the first LRR module. Residues in the boxed region are predicted to form a β-sheet/β-turn motif (xxLxLxx). Asterisks indicate invariant amino acids in AtPGIP1 and AtPGIP2. Dots represent gaps inserted in the sequences for the optimal alignment of LRR modules.