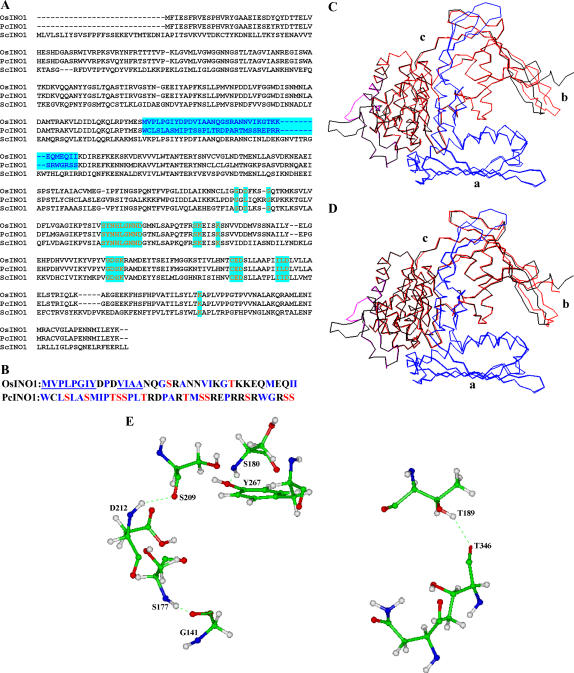
Figure 10.
Sequence alignment and modeling studies. A, Multiple sequence alignment of amino acids in OsINO1, PcINO1, and ScINO1. The 37-amino acid stretch is shown in blue. The conserved amino acid residues or stretches belonging to the core catalytic region are shown in red. B, Difference in the amino acid composition in the 37-amino acid residue stretch of OsINO1 and PcINO1. Hydrophobic residues of both OsINO1 and PcINO1 are marked blue. The hydrophobic stretches in OsINO1 are underlined. Ser and Thr residues are marked in red. C, Cα backbone superimposition of homology-modeled OsINO1 on yeast MIPS crystal. D, Cα backbone superimposition of homology-modeled PcINO1 on yeast MIPS crystal. The Cα backbone of yeast MIPS crystal is shown in black. The Cα backbones of OsINO1 and PcINO1 are shown in red. The 37-amino acid residue stretch is shown in cayan and the catalytic domain is shown in blue. a, Tetramerization/catalytic domain; b, central domain; and c, Rossmann fold domain. E, The four distinct hydrogen bond networks mediated by Ser and Thr in PcINO1 shown in ball-n-stick model.