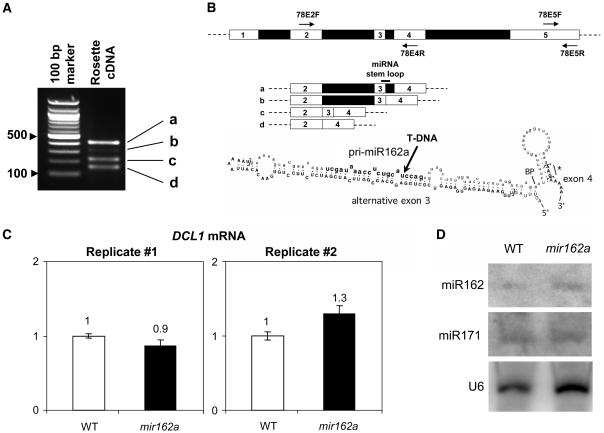
Figure 4.
Analysis of the npcRNA 78/MIR162a transcripts and molecular characterization of the mir162a mutant. A, RT-PCR detection of different npcRNA 78/MIR162a transcripts. RT-PCR was performed using npcRNA 78-specific primers located in exons 2 and 4 (78E2F/78E4R). B, Diagrammatic representation of the differentially spliced transcripts of npcRNA 78 as deduced from A and annotated structure of the miR162a predicted hairpin region. Exonic sequences are in uppercase bold letters, intronic sequences are in lowercase letters, and the miR162 sequence is in bold lowercase letters. Square brackets, 5′ and 3′ splice sites; BP, likely branch point; star, alternative codon. The arrow indicates the position of the SALK_107598 T-DNA insertion. C, Uncleaved DCL1 mRNA levels are similar in mir162a T-DNA insertion mutant and wild-type inflorescences. Real-time RT-PCR was performed using primers flanking the miR162-directed cleavage site in the DCL1 mRNA. Quantifications were normalized with ACTIN2. Values in wild-type inflorescences were arbitrarily fixed to 1. Quantifications were made in triplicate (error bars represent sds). Results for two biological replicates are presented. D, miR162 levels are comparable in wild-type and mir162a plants. Blots of RNA extracted from inflorescences of wild-type and mir162a plants were successively hybridized with miR162 and miR171 LNA-modified probes. Hybridization to a U6 probe served as a loading control.