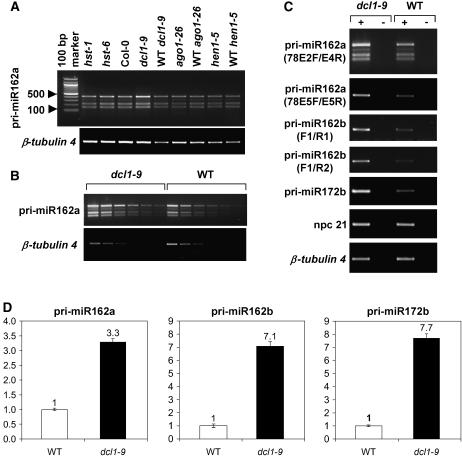
Figure 5.
npcRNA 78/MIR162a transcripts are stabilized in the dcl1-9 mutant. A, RT-PCR was performed using npcRNA 78-specific primers located in exons 2 and 4 (78E2F/78E4R) on RNA from rosettes of RNAi-related mutants. B, Dilution series of cDNAs from dcl1-9 and wild-type inflorescences shows consistently higher amplification by RT-PCR in the mutant when using the above-mentioned primers. C, Accumulation of miR162a and b primary transcripts in the dcl1-9 mutant. Specific primer pairs used for npcRNA 78/MIR162a amplification were located either in exons 2 and 4 (78E2F/78E4R) or in exon 5 (78E5F/78E5R). Primers pairs used for amplification of pri-miR162b (pri-miR162b-F1/R1 and pri-miR162b-F1/R2) did not reveal any alternative splicing for this transcript. The miR172b (EAT) primary transcript is a known pri-miRNA. npcRNA 21 is not affected by the dcl1 mutation. ±, RT-PCR performed with or without reverse transcriptase. D, Accumulation of miR162a and b primary transcripts in the dcl1-9 mutant measured by real-time RT-PCR. Primers used for npcRNA 78/MIR162a amplification were located in exon 5 (78E5F/78E5R). Primers pri-miR162b-F1 and R1 were used for amplification of pri-miR162b. Data were normalized with ACTIN2. Values in wild-type inflorescences were arbitrarily fixed to 1. Three technical replicates were performed for wild-type and dcl1-9 inflorescences. Values are means ± sds. For all these RT-PCR experiments, analyses were performed on homozygous mutants and their wild-type siblings.