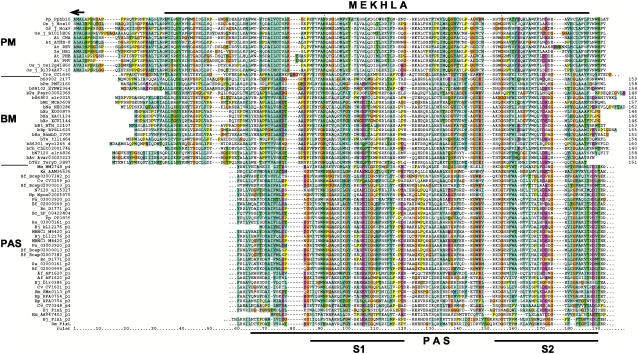
Figure 2.
Multiple sequence alignment of the extracted plant MEKHLA (PM) domains, the bacterial MEKHLA (BM) domain proteins, which are shown in their full length, and best-matching PAS domains extracted from bacterial PAS domain-containing proteins (PAS). The arrow at the top denotes the end of the HD-SAD domain in HD-ZIP III proteins. The extent of the MEKHLA domain is marked by a black line at the top; the extent of the PAS domain as defined by CDD at NCBI is marked by a black line at the bottom. Within the PAS domain, two better-conserved regions are referred to as S1 and S2 (Zhulin et al., 1997). We extracted the PAS domains of the best-scoring 20 proteins and added examples of FixL PAS domains for which the structure is known. The multiple sequence alignment is color coded according to the default of the ClustalX application. Dashes in the alignment indicate gaps, dots indicate omitted flanking sequences. For species abbreviations, see Table I; bacterial MEKHLA proteins are prefixed with a b.