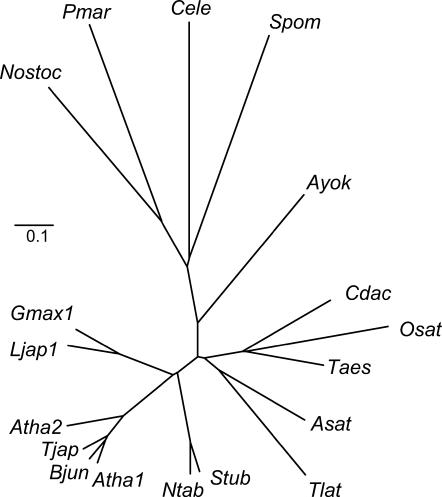
Figure 1.
Phylogenetic analysis of PCS proteins from cyanobacteria, nematodes, fungi, and plants. The unrooted tree was constructed using the neighbor-joining method (ClustalW) with 1,000 bootstrap replicates. Branch lengths are proportional to genetic distance, which is indicated by a bar (0.1 substitutions per site). GenBank accession numbers are as follows (in parentheses): Asat (Allium sativum, AAO13809), Atha1 (AtPCS1, AAD41794), Atha2 (AtPCS2, AAK94671), Ayok (Athyrium yokoscense, BAB64932), Bjun (Brassica juncea, CAC37692), Cdac (Cynodon dactylon, AAO13810), Cele (Caenorhabditis elegans, NP496475), Gmax (GmhPCS1, AAL78384), Ljap1 (LjPCS1, AY633847), Ntab (Nicotiana tabacum, AAO74500), Nostoc (Nostoc, NP485018), Osat (Oryza sativa, AAO13349), Pmar (Prochlorococcus marinus, NP894844), Taes (Triticum aestivum, AAD50592), Tjap (Thlaspi japonicum, BAB93119), Tlat (Typha latifolia, AAG22095), Spom (S. pombe, CAA92263), and Stub (Solanum tuberosum, CAD68109).