Figure 7.
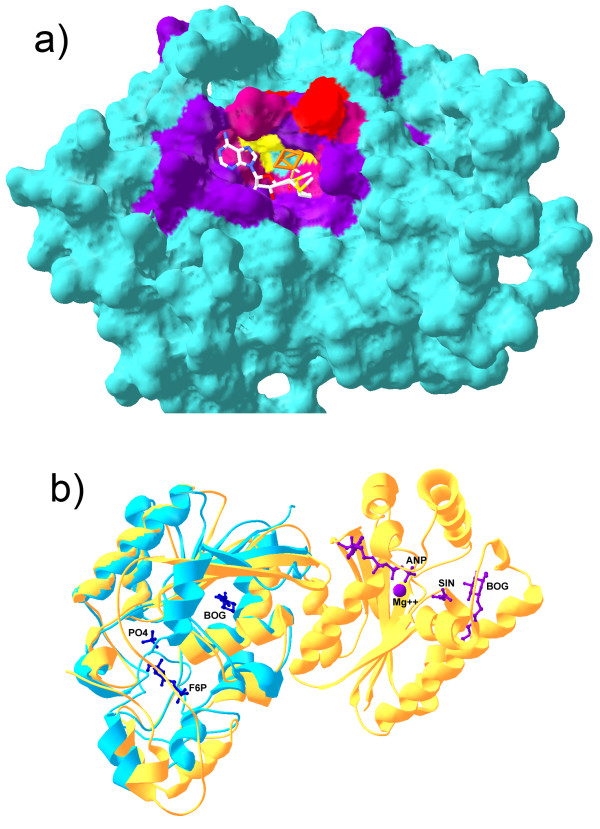
Binding sites predicted by SMID-BLAST. a) Shown is a comparative model of the predicted Elp3 domain of MiaB. The iron-sulfur cluster (orange) and SAM (CPK stick model) have had their co-ordinates transferred from the modelling template, PDB 1OLT chain A to illustrate how they might bind. The predicted Fe-S binding site residues are indicated in red, the predicted SAM binding residues are shown in purple, and the three cysteine residues which interact with the Fe-S cluster are indicated in yellow. A mixture of red and purple was used for residues common to both binding sites. b) Structural alignment of PDB 1RII chain A (phosphoglycerate mutase from M. tuberculosis, blue) and 2BIF chain A (6-phosphopructo-2-kinase/fructose-2,6-bisphosphatase from Rattus norvegicus, yellow). The small molecules from 2BIF are also shown along with their PDB short labels. Purple molecules associate with the N-terminal domain of 2BIF chain A, while blue molecules associate with the C-temrinal domain. Note that BOG was part of the crystallization buffer in this example. Structures were aligned with Swiss PDBViewer.
