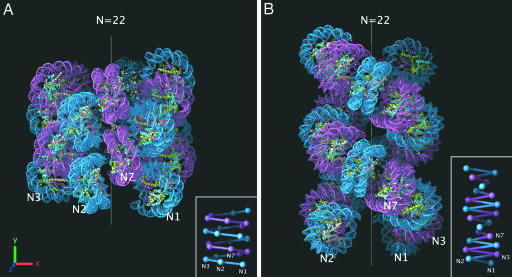
Fig. 3.
Modeling the 30-nm chromatin fiber. (A) Interdigitated one-start helix model built by using the constraints imposed by our measurements of diameter and nucleosome packing ratio (Fig. 2). The helix contains 22 nucleosomes and has a diameter of 33 nm and height of ≈33 nm. Alternate helical gyres are colored marine and magenta. (B) Two-start helical crossed linker model adapted from the idealized model reported by Schalch et al. (38). The model maintains the same parameters for rise per nucleosome and azimuthal rotation angle while extrapolating to a 177-bp nucleosome repeat length by increasing the radius by 3.4 nm (roughly approximating the addition of 10 bp of extra linker DNA lying orthogonal to the fiber axis). The resulting helix contains 22 nucleosomes and has a 28.4-nm diameter and ≈47-nm height. Alternate nucleosome pairs are colored marine and magenta. The positions of the first, second, third, and seventh nucleosomes in the linear DNA sequence are marked on both models with N1, N2, N3, and N7. (Insets) Schematic representations of both atomic models showing the proposed DNA connectivity.