Figure 6.
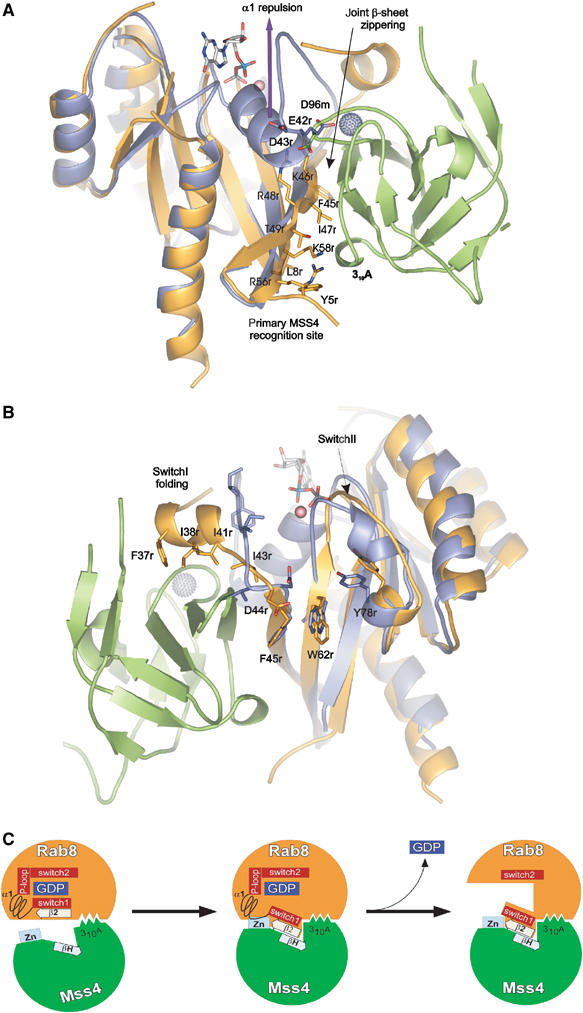
Structural rearrangement of Rab structure upon MSS4 binding. Rab8 and Sec4:GDP:Mg2+ are superimposed. The Rab8 molecule is shown in orange, Sec4 in violet, GDP and Mg2+ in ball and stick representation, MSS4 in green, and the MSS4-bound Zn2+ ion as a gray ball. See the section on Rab8:MSS4 complex formation and nucleotide release for details. Panels (A) and (B) are given in 180° rotation. (C) Model for MSS4-catalyzed nucleotide displacement from Rab8:GDP. In the first step, Rab8:GDP (orange) (Tyr5, Leu8, Phe45, Ile47, Lys58) associates loosely with the 310A helix (Met74, Phe75, and Ile76) of MSS4 (green). The complex association is followed by formation of a shared intermolecular β-sheet (indicated by the boxed arrows) between β2 of Rab8 and βH of MSS4. As Switch 1 and β2 are directly adjacent in the sequence, the movement of β2 is accompanied by a shift of Switch 1 towards the surface of MSS4. Due to these movements, MSS4 and Rab8 form a tight complex in which the Zn-finger (indicated in light blue) region of MSS4 would clash sterically with α1 of Rab8. Hence, α1 is displaced and unfolds together with the P-loop, promoting nucleotide release. The disordered nucleotide-binding pocket of Rab8 will not allow nucleotide binding unless α1 und the P-loop are refolded.
