Figure 5.
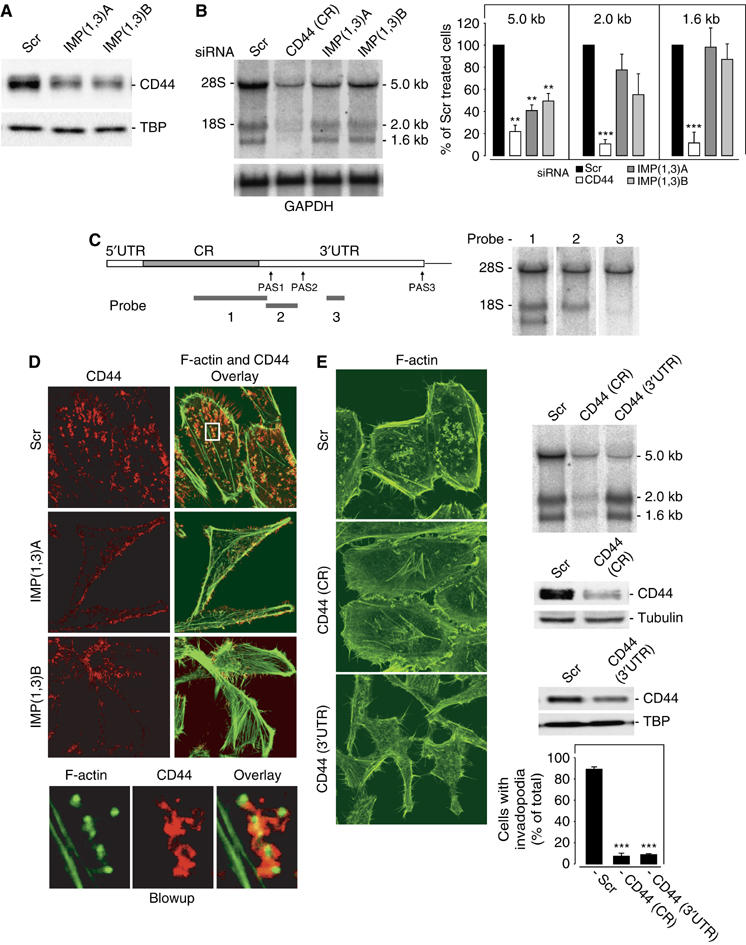
CD44 in invadopodia formation. CD44 protein and mRNA levels in IMP-depleted cells. (A) Western blot analysis of CD44 in HeLa cells transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs, respectively. TBP was used as loading control (B) Northern blot analysis of CD44 mRNA under the same conditions (left panel). The right panel shows the quantification of the 5.0, 2.0 and 1.6 kb CD44 transcripts during knockdown. GAPDH was used as loading control. (C) Schematic representation of the full-length CD44 transcript outlining the position of the different polyadenylation signals (PAS) and probes designated 1, 2 and 3, that were used in the Northern blot analysis shown to the right. 5′UTR—5′untranslated region; CR—coding region; 3′UTR—3′untranslated region. (D) CD44 is associated with invadopodia formation. HeLa cells were stained with mouse anti-CD44 antibody and phalloidin to depict F-actin. The blowup below shows that the CD44 protein is located in close proximity to invadopodia. (E) Loss of invadopodia in CD44-depleted cells. HeLa cells were treated with Scr siRNA or siRNA targeting the CD44 mRNA coding region (CR) or the 3′UTR (left panel). The upper right panel shows a Northern blot analysis of the siRNA-treated cells. At 72 h after transfection, the cells were harvested and the level of CD44 protein was determined by Western blot analysis (middle right panel). β-Tubulin or TBP was included as loading control. Finally, the cells were stained for F-actin and the number of invadopodia were counted (lower right panel). Data are presented as the mean value±s.d. of three independent experiments (lower right panel). Asterisks depict statistically significant differences between the Scr and CD44 siRNA-treated cells, by an unpaired, unequal t-test (***P<0.001).
