Abstract
Coronaviruses are the causative agents of respiratory disease in humans and animals, including severe acute respiratory syndrome. Fusion of coronaviruses is generally thought to occur at neutral pH, although there is also evidence for a role of acidic endosomes during entry of a variety of coronaviruses. Therefore, the molecular basis of coronavirus fusion during entry into host cells remains incompletely defined. Here, we examined coronavirus-cell fusion and entry employing the avian coronavirus infectious bronchitis virus (IBV). Virus entry into cells was inhibited by acidotropic bases and by other inhibitors of pH-dependent endocytosis. We carried out fluorescence-dequenching fusion assays of R18-labeled virions and show that for IBV, coronavirus-cell fusion occurs in a low-pH-dependent manner, with a half-maximal rate of fusion occurring at pH 5.5. Fusion was reduced, but still occurred, at lower temperatures (20°C). We observed no effect of inhibitors of endosomal proteases on the fusion event. These data are the first direct measure of virus-cell fusion for any coronavirus and demonstrate that the coronavirus IBV employs a direct, low-pH-dependent virus-cell fusion activation reaction. We further show that IBV was not inactivated, and fusion was unaffected, by prior exposure to pH 5.0 buffer. Virions also showed evidence of reversible conformational changes in their surface proteins, indicating that aspects of the fusion reaction may be reversible in nature.
For all enveloped viruses, a critical event during entry into cells is the fusion of the viral envelope with the membrane of the host cell (13). Our current understanding of viral fusion has been driven by fundamental problems first solved with influenza hemagglutinin (HA) (50). Whereas the trigger for HA-mediated fusion is the low pH of the endosome, other viruses (e.g., paramyxoviruses and most retroviruses) undergo a receptor-primed fusion with the plasma membrane at neutral pH (13).
Coronaviruses (CoV) have recently received much attention due to the outbreak of severe acute respiratory syndrome (SARS) (22, 28), but there is little consensus as to whether coronavirus entry and fusion occur following endocytosis or at the plasma membrane (6, 16, 21, 43). Coronaviruses are enveloped positive-strand RNA viruses that replicate in the cytoplasm (28). They have a distinctive set of club-shaped spikes on their envelope, and the spike protein (S) is the primary determinant of cell tropism and pathogenesis, being responsible (and apparently sufficient) for receptor binding and fusion (16). However, other envelope proteins are present: the M protein, the E protein, and (in some coronaviruses) an HE protein (28). The coronavirus S protein is categorized as a class I fusion protein, based on the presence of characteristic heptad repeats (3, 9, 26); as such, it shows features of the fusion proteins of influenza virus (HA), retroviruses (Env), and paramyxoviruses (F and HN), for which there is extensive characterization at the structural and biophysical levels (11).
Although class I fusion proteins share similar structural features, they can have quite different biological properties, i.e., they can be triggered for fusion by low pH or by coreceptor interaction. Influenza virus is a classic example of low-pH-induced fusion (50), and retroviruses, such as human immunodeficiency virus (HIV), are well-characterized systems in which coreceptor interaction triggers the necessary conformational changes in Env that allow fusion to occur (14). In the case of coronaviruses, receptor-induced conformational changes have been described (29, 34, 57), and the fusions of murine coronavirus, bovine coronavirus, and infectious bronchitis virus (IBV) are considered to exhibit a near-neutral or slightly alkaline pH optimum (30, 41, 51, 54). Since these fusion data are exclusively based on cell-cell fusion assays with S-expressing cells, the nature of coronavirus fusion during entry into host cells remains incompletely defined.
To understand the molecular details of coronavirus fusion, biochemical and biophysical studies are needed. However, a significant problem for most coronaviruses is the fact that the virus is difficult to purify for such studies. Because of this, direct virus-cell fusion assays have not been performed for any coronavirus, and the molecular basis of fusion during virus entry remains elusive. Here, we examined coronavirus-cell fusion using fluorescence-dequenching (FdQ) assays (18) of octadecyl rhodamine (R18)-labeled viruses with host cells, using IBV, a coronavirus that can be isolated, purified, and labeled appropriately for FdQ studies. Our IBV model, for the first time, allows FdQ studies of coronavirus-cell fusion to be performed. We show that fusion does not occur at neutral pH and that fusion activation is a direct low-pH-dependent process occurring within acidic endosomes, with a half-maximal rate of fusion at pH 5.5.
MATERIALS AND METHODS
Viruses, cells, and infections.
IBV (strains Beaudette and Massachusetts 41 [M41]; provided by Benjamin Lucio-Martinez, Unit of Avian Health, Cornell University), influenza virus (strain A/WSN/33), and Sendai virus (strain Cantell; American Type Culture Collection [ATCC]) were propagated in 11-day-old embryonated specific-pathogen-free chicken eggs. Vesicular stomatitis virus (VSV, strain Orsay; ATCC) was propagated in BHK cells. The BHK cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (Cellgro) containing 10% fetal bovine serum and passaged twice weekly. Primary chicken kidney (CK) cells were isolated from the kidneys of 11- to 14-day-old chickens and seeded in medium 199 (Cellgro) containing 5% fetal bovine serum.
IBV was purified using a 30 to 65% (vol/vol) sucrose step gradient. Specific-pathogen-free chicken eggs were infected with IBV (approximately 102 50% embryo infectious doses/egg), and the allantoic fluid was harvested at 24 h (IBV Beaudette) or 48 h (IBV M41) postinoculation. The allantoic fluid was clarified by centrifugation at 2,000 × g for 15 min and then centrifuged at 18,000 rpm in an SW32 rotor (Sorvall). The pellet was loaded onto a 30 to 65% (vol/vol) sucrose step gradient and centrifuged at 25,000 rpm in an SW32 rotor. The virus-containing band at the 65% sucrose interface was concentrated using an SW32 rotor at 18,000 rpm. The pellet was gently rinsed with cold phosphate-buffered saline (PBS) to remove residual sucrose, and purified sucrose-free virus was then resuspended in 2-mg/ml stocks in PBS.
Infections were performed essentially as described previously (47). Briefly, viral stocks were diluted in binding medium (RPMI 1640 medium containing 0.2% bovine serum albumin [BSA], pH 7.4), and unless otherwise described, virus was adsorbed for 60 min at 37°C. The cells were then maintained in growth medium containing 2% fetal bovine serum at 37°C before fixation and analysis. NH4Cl, bafilomycin A1, monensin, and E64-d were obtained from Calbiochem.
Virus-cell fusion assay.
Fusion assays were based on fluorescence dequenching of R18-labeled virus (18). Typically, 100 μl of purified virus (2 mg/ml) was labeled by the addition of 2.5 μl of 1.7 mM R18 (Molecular Probes), and the mixture was incubated in the dark on a rotary shaker at room temperature for 60 min. Excess dye was removed with a Sephadex G25 column (Pharmacia). Fifteen microliters of labeled virus (approximately 5 PFU/cell) was bound to 1.5 × 106 cells at 4°C for 1 h in binding buffer. Unbound virus was removed by washing it with binding buffer, and the cells were resuspended in fusion buffer (5 mM HEPES, 5 mM MES [morpholineethanesulfonic acid], 5 mM succinate, 150 mM NaCl buffer, pH 7.0, 15 μM monensin) at 37°C. To examine the fusion of IBV with the cell membrane, fusion was triggered by adding a predetermined amount of 250 mM HCl to obtain a final pH between 5.0 and 7.0. Control viruses were similarly labeled and induced to fuse at pH 7.0 (Sendai virus) (42), pH 5.0 (influenza virus) (20), or pH 5.5 (VSV) (44). Fluorescence dequenching was measured using a QM-6SE spectrofluorimeter (Photon Technology International), with excitation and emission wavelengths (λEx and λEm) set to 560 nm and 590 nm, respectively. Fusion efficiency was determined following the addition of Triton X-100 (final concentration, 1%) to obtain 100% dequenching.
Immunofluorescence microscopy.
Immunofluorescence microscopy was essentially performed as described previously (47), except with methanol fixation for IBV. IBV was identified using anti-S1 monoclonal antibody 15:88 (25). Influenza virus was detected using mouse monoclonal antibody H16 L10 4R5 (anti-NP) (ATCC). Sendai virus was identified with a chicken polyclonal anti-Sendai virus antibody (U.S. Biological), and VSV was detected with mouse monoclonal antibody P5D4 (Roche Applied Science). The secondary antibodies used were Alexa 488-labeled or Alexa 568-labeled goat anti-mouse or anti-chicken immunoglobulin G (Molecular Probes). Cells were viewed on a Nikon Eclipse E600 fluorescence microscope, and images were captured with a Sensicam EM camera and IPLab software before transfer into Adobe Photoshop 7 and determination of infection frequency.
bis-ANS fluorescence.
1,1′-bi(4-anilino)naphthalene-5-5′-disulfonic acid (bis-ANS) was obtained from Molecular Probes. Purified IBV (Beaudette), influenza virus (A/WSN/33), or VSV (Orsay) (2 mg/ml) was incubated at pH 7.0 or pH 5.0 or was treated with pH 5.0 fusion buffer for 5 min before neutralization to pH 7.0. bis-ANS (1 mM) was then added, and the fluorescence intensity was measured after 5 min (λEx = 400 nm; λEm = 490 nm) using a Molecular Devices Spectramax Gemini XS fluorimeter.
ELISAs.
Enzyme-linked immunosorbent assay (ELISA) plates were coated with 100 ng of virus protein at 4°C for 12 h and washed twice with 0.5% BSA-PBS. The wells were blocked with 0.5% BSA-PBS at 4°C for 1 h, and the plates were incubated with mouse monoclonal anti-S1 (15:88) or S2 (9:4) antibody at room temperature. The wells were then blocked with 0.5% BSA-PBS and incubated with anti-mouse horseradish peroxidase at room temperature for 1 h. The wells were developed with ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] and analyzed at a λ of 405 nm.
RESULTS
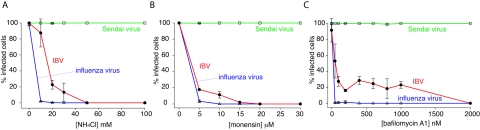
Studies of IBV infection are typically performed in primary chicken cells or embryonated eggs due to the restricted tropism of the virus (8), and the IBV strain Beaudette has been adapted to efficiently replicate in, and rapidly kill, chicken embryos (1, 15, 36). In addition to its embryo-lethal phenotype, IBV Beaudette has acquired extended species tropism and infects a variety of continuous cell lines, including baby hamster kidney (BHK) cells (40); the Beaudette strain therefore serves as an excellent cell culture model for IBV infection. To examine the functional role of a low-pH-dependent fusion trigger for coronavirus infection, we first carried out infections of IBV Beaudette in BHK cells in the presence of acidotropic bases and drugs that neutralize the low pH of endosomes, using influenza virus and Sendai virus as controls. When added prior to virus binding, the addition of ammonium chloride resulted in a dose-dependent inhibition of coronavirus infection but had no effect on Sendai virus (Fig. 1A). Slightly higher levels of ammonium chloride were necessary to completely block IBV entry than were necessary with influenza virus. A similar situation was found with the ionophore monensin; IBV infection was inhibited at slightly higher drug concentrations than for influenza virus, and Sendai virus was unaffected (Fig. 1B). We also examined infection in the presence of bafilomycin A1 (BafA), a potent inhibitor of the vacuolar H+-ATPase. In this case, influenza virus was very sensitive to BafA treatment (complete inhibition of infection at 20 nM), and Sendai virus was unaffected. IBV was sensitive to BafA in a dose-dependent manner, although much higher concentrations of the drug (approximately 1 μM) were necessary to achieve complete inhibition of infection; however, levels as low as 100 nM gave approximately 80% inhibition (Fig. 1C). The reasons for this are unclear. To ensure that the effects of the above-mentioned compounds were specific to virus entry, we also performed experiments in which the inhibitor was added 60 min after initiation of infection. In that case, viruses were expected to have exited the endosomal compartments and entered the cytosol within 60 min and were therefore resistant to endosome inhibitors. For ammonium chloride, BafA, and monensin, infection levels were similar to those of untreated controls when added after 60 min (not shown), showing that low endosomal pH is specifically required for coronavirus entry.
FIG. 1.
Infection of IBV is prevented by treatment of cells with inhibitors of endosome acidification. BHK cells were treated with various concentrations of NH4Cl (A), monensin (B), or bafilomycin A1 (C) for 30 min and infected with IBV strain Beaudette, influenza virus strain A/WSN/33, or Sendai virus strain Cantell at a multiplicity of infection of 1 to 5 infectious units/cell. Infectivity was determined by immunofluorescence microscopy 8 h postinfection using anti-IBV S1 monoclonal antibody (15:88), anti-influenza NP monoclonal antibody H16 L10 4R5, or chicken anti-Sendai virus antibody. For quantification, >300 cells were scored in three independent experiments. The error bars represent the standard errors of the mean.
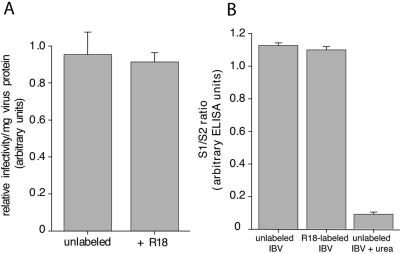
To directly examine the coronavirus-cell fusion event, we purified IBV strain Beaudette from the allantoic cavities of embryonated chicken eggs and labeled virions with the lipophilic fluorescent dye R18. The virions were labeled with a sufficiently high concentration of R18 to achieve self-quenching of the probe, and such R18-labeled virus retained high levels of specific infectivity (Fig. 2A). We also examined the integrity of our R18-labeled virions, as coronavirus entry experiments have been considered technically difficult to study, in part because of the tendency of the S1 component of the spike protein to detach from the virions (23). We monitored the relative ratio of S1 and S2 by ELISA, using specific monoclonal antibodies recognizing these domains (25). No loss of S1 was seen after R18 labeling and subsequent purification (Fig. 2B); however, S1 was efficiently released by urea, a treatment known to detach the S1 domain (7).
FIG. 2.
R18-labeled IBV virions retain infectivity in BHK cells and show no loss of S1. To determine relative infectivity, purified IBV strain Beaudette was incubated with or without R18. The protein concentration of each virus preparation was assayed via Bio-Rad protein assay, and virus infectivity was assessed by infecting BHK cells using immunofluorescence microscopy (A). To determine virus integrity, 500 ng of either R18-labeled or unlabeled IBV strain Beaudette was use to coat an ELISA plate, and the S1/S2 ratio of each sample was determined via anti-S1 (15:88) and anti-S2 (9:4) monoclonal antibody staining, followed by anti-mouse horseradish peroxidase labeling and ABTS development (B). The error bars represent the standard errors of the mean.
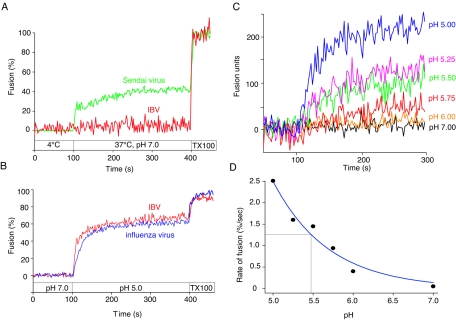
For fluorescence-dequenching studies, labeled virus was bound to the surfaces of BHK cells at 4°C and shifted to 37°C in fusion buffer (pH 7.0) in the presence of monensin to prevent any entry from acidic endosomes. Upon lipid mixing that occurs during membrane fusion, the quenched probe is diluted in the cell membrane and undergoes dequenching, which can be monitored as an increase in fluorescence signal by spectrofluorimetry. Even after a significant time at 37°C (300 s), we saw little or no dequenching of virus signal that would indicate virus-cell fusion at neutral pH (Fig. 3A). However, addition of Triton X-100, caused extensive dequenching, showing that the virus binding had occurred and the virions were labeled appropriately. This indicated that the lack of dequenching was due to a lack of fusion activity at pH 7.0. In contrast, Sendai virus (a paramyxovirus well established to fuse at neutral pH) (42) gave extensive dequenching under the same conditions (Fig. 3A).
FIG. 3.
Fusion of IBV with host cells is low pH dependent. R18-labeled IBV strain Beaudette or Sendai virus strain Cantell was bound to BHK cells at 4°C for 60 min and then injected into a spectrofluorimeter cuvette containing 1 ml of pH 7.0 buffer at 37°C (t = 100 s) (A). Samples were monitored for fluorescence dequenching at 37°C for 300 seconds before addition of 1% Triton X-100 (final concentration) to obtain complete (100%) dequenching. (B) Similarly, R18-labeled IBV strain Beaudette or influenza virus strain A/WSN/33 was bound to BHK cells at 4°C, but samples were added to pH 7.0 buffer at 37°C (t = 0 s). At t = 100 s, the buffer pH was reduced to 5.0 and samples were monitored for fluorescence dequenching at 37°C. At t = 400 s, the final concentration of 1% Triton X-100 was added to obtain 100% dequenching. (C) Samples were treated as described for panel B under various pH conditions, and dequenching activities are shown in terms of actual fusion units. The initial rate of fusion obtained from panel C was analyzed by four-parameter exponential decay and is plotted against various pHs (D). The pH which gave the half-maximal initial rate of IBV fusion (pH1/2) is indicated.
As IBV appeared to be unable to fuse with cells at neutral pH, we wished to determine if coronavirus fusion was pH dependent. We first examined influenza virus (an orthomyxovirus well-established to fuse at pH 5.0 to 5.5) (50). As expected, influenza virus showed no fusion activity at pH 7.0 in the presence of 15 μM monensin and gave extensive cell surface dequenching when the pH was lowered to 5.0 (Fig. 3B). When we examined the coronavirus IBV, we saw that fusion was similarly sensitive to low pH (Fig. 3B). At pH 5.0, we obtained extensive dequenching, with the overall extent of fusion between 40 and 60% of that in the presence of Triton X-100 (the addition of which results in complete dequenching), with little or no appreciable lag time after pH change. At pH 5.5, high levels of dequenching were still observed (Fig. 3C), and limited dequenching could still occur at pH 5.75; however, at pH 6.0 and above, dequenching was negligible. Below pH 5.0, the IBV fusion reaction was unstable and calibration was not possible (V. C. Chu and G. R. Whittaker, unpublished results).
To define a pH threshold for fusion, we calculated the initial rate of fusion between pH 7.0 and 5.0 (Fig. 3D). Typically, we did not see an abrupt threshold for low-pH-activated IBV fusion, as would be expected for influenza virus (20), but a more gradual increase in fusion activity between pH 6.0 and 5.0. In our FdQ system, the half-maximal pH (pH1/2) at which IBV fusion occurred was 5.5.
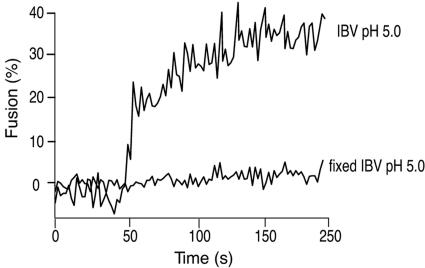
One potential problem that has been reported for R18-based fluorescence-dequenching assays is due to nonspecific probe transfer when virus is bound to the surfaces of cells (39). However, in our assays, we see only very limited probe transfer, and experiments using viruses pretreated with a low level of paraformaldehyde and exposed to low pH show no significant dequenching (Fig. 4). Therefore, we are confident that our dequenching results measure bona fide lipid mixing due to virus fusion and are not due to nonspecific probe transfer.
FIG. 4.
R18-labeled IBV does not undergo significant nonspecific dye transfer. R18-labeled IBV strain Beaudette was untreated or pretreated with 0.5% paraformaldehyde (fixed IBV) before binding to BHK cells. An FdQ assay was performed on each sample as described for Fig. 4B. The pH was reduced to 5.0 to induce fusion at t = 50 s.
IBV strain Beaudette is a laboratory-adapted strain that is nonpathogenic in adult animals. Consequently, we wished to examine whether low-pH dependence for coronavirus fusion was also necessary in a more clinically relevant system. We purified and labeled virions of the M41 serotype of IBV, which are common causes of disease outbreaks (8). R18-labeled M41 virus was subjected to FdQ assays with CK cells and showed a pattern of pH dependence similar to that of the Beaudette strain examined previously, with an essentially identical pH1/2 of 5.5 (not shown).
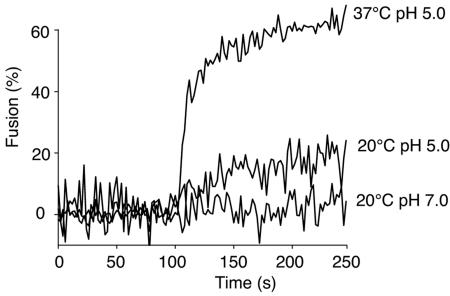
One feature of viruses that fuse at low pH that is not shared by pH-neutral viruses is that fusion can still occur at lower temperatures (13). Because of this, we examined IBV fusion at 20°C. After fusion was induced by the addition of pH 5.0 buffer, we observed limited but significant dequenching (Fig. 5). The initial rate of fusion at 20°C was approximately 25% of the value obtained at 37°C. These data indicate that the coronavirus IBV fits into the general pattern of pH-dependent fusion (i.e., influenza virus-like), rather than pH-independent fusion (i.e., retrovirus- or paramyxovirus-like).
FIG. 5.
Limited but significant IBV fusion occurs at lower temperature. R18-labeled IBV strain Beaudette was bound to BHK cells at 4°C for 60 min and then added to a spectrofluorimeter cuvette at either 20°C or 37°C (t = 0 s). The pH was reduced to 5.0 at t = 100 s or maintained at 7.0 while FdQ activity was monitored.
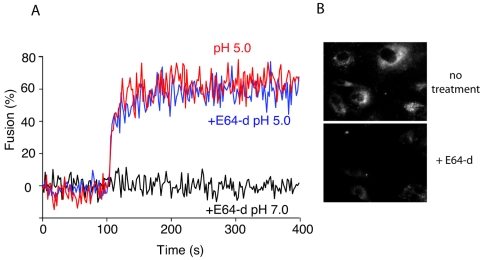
To accommodate low-pH-dependent entry of SARS-CoV with a proposed pH-neutral fusion reaction, it has recently been suggested that the low pH of the endosome may act indirectly, possibly activating an endosomal protease (cathepsin L) necessary for S protein cleavage and subsequent fusion activation (35, 48). Although our fluorescence-dequenching experiments clearly showed a direct low-pH-dependent activation of IBV fusion, it remained possible that by dropping the external pH in our assay we were activating a secreted or cell surface-bound protease that was then responsible for fusion activation. To examine this possibility, we repeated our fluorescence-dequenching assays in the presence of the cathepsin-sensitive protease inhibitor E64-d (Fig. 6). We observed no detectable difference in fusion activation in the presence or absence of E64-d (Fig. 6A), whereas a fluorescence assay of cathepsin L activity showed marked reduction of enzyme activity in control BHK cells (Fig. 6B). Quantification of the pixel intensity in Fig. 6B showed >85% inhibition of cathepsin L activity after E64-d treatment (data not shown). These data confirm that IBV fusion activation is occurring in a direct low-pH-dependent manner.
FIG. 6.
Cysteine proteases are not essential for IBV fusion activation during viral entry. BHK cells were pretreated with 400 μg/ml of E64-d, and R18-labeled IBV strain Beaudette bound at 4°C for 60 min. An FdQ assay was performed as described for Fig. 4B in the presence or absence of E64-d throughout the entire experiment. Virus-cell fusion was triggered by reducing the buffer pH to 5.0 at t = 100 s. In panel B, E64-d activity was assessed using a Cathepsin L Activity Detection Kit (Calbiochem) in the presence or absence of drug treatment, according to the manufacturer's instructions.
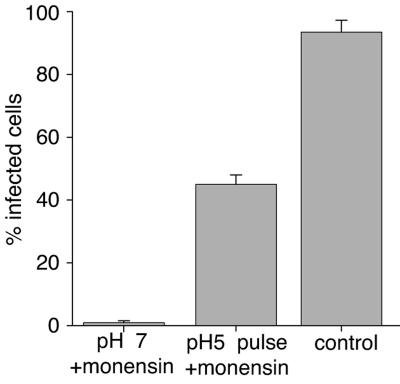
Fluorescence-dequenching assays such as the one employed here are lipid-mixing assays, and probe dequenching would still be apparent if fusion were arrested at the hemifusion stage. To ensure that the low-pH-induced fusion event we measured by fluorescence dequenching resulted in fusion progression to later stages, i.e., fusion pore formation and expansion, we carried out a content-mixing assay. We bound IBV to the surfaces of cells and assayed the delivery of the virus genome (and subsequent viral replication in the cytoplasm) when fusion was induced at the cell surface. To ensure that we were monitoring only fusion from the cell surface, we neutralized the low endosomal pH with 15 μM monensin as a control. When the external medium was maintained at pH 7, we saw an almost complete block of virus infection, indicating entry through acidic endosomes (Fig. 7). In contrast, when the surface-bound virus was exposed to pH 5.0 buffer for 2 min, IBV efficiently infected cells by fusing with the plasma membrane, albeit at somewhat reduced levels compared to that occurring through the normal entry pathway, i.e., through endosomes (Fig. 1). These data are in general agreement with other enveloped viruses that enter cells by pH-dependent endocytosis (33) and confirm that low pH induces complete fusion of the IBV S protein with host cells.
FIG. 7.
Content mixing occurs following low-pH-induced IBV fusion at the cell surface. IBV strain Beaudette (multiplicity of infection, 5 infectious units/cell) was bound to the surfaces of BHK cells at 4°C for 60 min and was treated with 15 μM monensin to block virus entry from endosomes or was left untreated as a control. In the pH 5 pulse sample, the buffer pH was reduced to 5.0 in the presence of 15 μM of monensin for 2 min at 37°C and then replaced with 2% Dulbecco's modified Eagle's medium with monensin at 37°C for 8 h. Genome delivery and viral replication were monitored by expression of S glycoprotein via immunofluorescence microscopy. For quantification, >100 cells were scored in three independent experiments. The error bars represent the standard deviations of the mean.
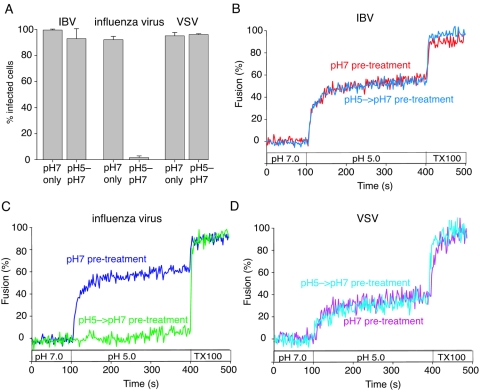
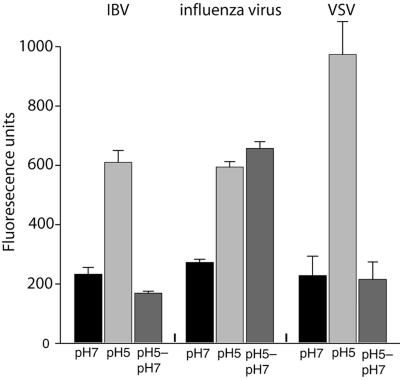
Many low-pH-dependent viruses (e.g., influenza virus) undergo essentially irreversible conformational changes that trigger fusion; hence, exposure of the virus to low pH and subsequent neutralization prior to exposure to cells efficiently inactivates virions and abolishes infection (4). However a few viruses (notably VSV) have reversible triggers for fusion and are not inactivated by low-pH exposure and subsequent neutralization (17). To examine the possible reversibility of coronavirus fusion, we exposed IBV (Beaudette) to pH 5.0 buffer for 10 min and subsequently reneutralized the virus to pH 7.0 before infection of BHK cells. Whereas influenza virus infection was completely abrogated by pretreatment with pH 5.0 buffer, VSV was essentially unaffected. Infection with the coronavirus IBV was almost completely resistant to low-pH exposure and subsequent neutralization of virions (Fig. 8A). We also performed fluorescence-dequenching assays of IBV that had been pretreated with pH 5.0 buffer, which showed no discernible differences from IBV that was not pretreated (Fig. 8B). As expected, influenza virus fusion was abolished by pH 5.0 pretreatment (Fig. 8C), and VSV fusion was insensitive to pH 5.0 pretreatment (Fig. 8D).
FIG. 8.
Infection and fusion by IBV are not prevented by pretreatment of virions with low-pH buffer. IBV strain Beaudette, influenza virus strain A/WSN/33, and VSV strain Orsay were purified and incubated in pH 5.0 buffer for 10 min before neutralization to pH 7.0 (pH 5-pH 7) or were maintained at pH 7.0 (pH 7 only) (A). BHK cells were then infected with virus at a multiplicity of infection of 5 infectious units/cell, and infection was monitored by immunofluorescence microscopy with monoclonal antibodies anti-IBV S1 (15:88), anti-influenza virus NP H16 L10 4R5, and anti-VSV G (P5D4) after 8 h of incubation. For quantification, >100 cells were scored in three independent experiments. The error bars represent the standard deviations of the mean. (B, C, and D) IBV strain Beaudette, influenza virus strain A/WSN/33, and VSV strain Orsay were treated with either pH 5.0 or 7.0 buffer for 10 min before neutralization and then bound to BHK cells at 4°C for 60 min. FdQ assays were then performed and monitored as described for Fig. 3B, and buffer pH was reduced from pH 7.0 to 5.0 at t = 100 s.
To investigate more directly the possible reversible nature of low-pH-dependent conformational changes that occur for IBV, we employed the fluorophore bis-ANS. This probe is sensitive to the polarity of its environment, so that it is virtually nonfluorescent in aqueous solution but becomes strongly fluorescent when it is bound to hydrophobic sites in proteins (46). bis-ANS has been widely used as a probe to monitor protein folding and unfolding and can be used either on purified protein or on intact virions (27). We incubated purified IBV with bis-ANS at pH 7 or pH 5 and saw a pronounced increase in bis-ANS binding at the lower pH, indicating exposure of hydrophobic domains or residues of the S protein (Fig. 9). When the pH was returned to neutral, the bis-ANS fluorescence returned to background levels, indicting a reversion of S protein conformational changes. In contrast, influenza virus (which possesses an irreversible fusion trigger) maintained its high levels of bis-ANS binding when pH 5-treated virus was neutralized. VSV also showed an increase in bis-ANS fluorescence at pH 5, followed by return to baseline levels upon reneutralization, indicative of the reversible conformational changes occurring in the VSV G protein. Overall, these data suggest that the low-pH-activated conformational changes are required prior to IBV coronavirus fusion and that these conformational changes may be reversible in nature.
FIG. 9.
bis-ANS labeling demonstrates S glycoprotein conformation reversibility. Purified IBV strain Beaudette, influenza virus strain A/WSN/33, and VSV strain Orsay were pretreated with pH 7.0, pH 5.0, or pH 5.0 buffer followed by neutralization to pH 7.0 at 37°C for 10 min. Then, each sample was subjected to bis-ANS binding at 37°C for 5 min before analysis by fluorimetry. The samples represent the means of five replicate wells, and the error bars represent the standard deviations of the mean.
DISCUSSION
Using an established assay of virus-cell fusion, we show here that fusion of the coronavirus IBV with host cells does not occur at neutral pH and that fusion activation is a low-pH-dependent process, with a half-maximal rate of fusion at pH 5.5. Little or no fusion occurred above a pH of 6.0. The pH optimum for fusion was 5.0 at 37°C, where fusion occurred rapidly, reaching maximal extent (approximately 40%) within 60 s. Fusion still occurred at lower temperatures (e.g., 20°C), albeit with reduced kinetics and extent. As such, the coronavirus IBV shows many similarities to pH-dependent viruses, such as influenza virus or VSV, and little or no similarity to retroviruses or paramyxoviruses, which fuse at neutral pH.
In general, coronaviruses have been categorized as undergoing pH-independent fusion (13). How, then, can we rationalize the substantial data showing a neutral-pH coronavirus fusion reaction (at least for cell-cell fusion) with our own data that clearly show activation of virus-cell fusion at pH 5.5? It is possible that cell-cell fusion assays involve overexpression of the viral S protein and its receptor, which might provide an environment where even highly inefficient membrane fusion events (i.e., those occurring at neutral pH) might be visualized. Alternatively, an explanation for the apparently discrepant data is that the IBV S protein undergoes reversible conformational changes that might account for fusion activation, as is the case for VSV (17). Reversibility in fusion activation may account for the ability of IBV to be highly syncytial yet have a low-pH-dependent fusion trigger during virus fusion. With VSV infection, syncytia can be observed even with a fusion protein that is clearly triggered by low pH (45). In a similar fashion, a fraction of the IBV S protein that is expressed at the cell surface (32, 53) may transiently attain a fusion-competent state during maturation and delivery, allowing some degree of cell-cell fusion at the plasma membrane. While the pH of the Golgi is only mildly acidic, the pH of secretory vesicles can be as low as 5.5 (56) and would be low enough to activate fusion. In support of this model, brief treatment of IBV-infected cells with pH 5.0 buffer leads to an approximately 50% increase in syncytium formation (data not shown).
Unlike viruses with well-characterized class I fusion proteins (e.g., retroviruses and influenza virus), coronaviruses do not have ubiquitously cleaved spike proteins. It has been suggested that cleavage of the S protein might enable pH-independent, receptor-primed triggering of fusion activity in the case of coronaviruses (21, 49). However, it is noteworthy that the IBV S protein is found in a completely cleaved S1/S2 form (data not shown), and so in this case there is no correlation between cleavage and the acquisition of pH-independent triggering of fusion. Although the coronavirus S proteins clearly have a highly helical secondary structure and heptad repeat domains (3), the lack of a requirement for proteolytic activation, combined with the reversible nature of fusion activation, suggests that S protein does not behave biologically as a canonical class I fusion protein. Several coronaviruses, including IBV and porcine transmissible enteritis virus, are clearly sensitive to endosome acidification during entry into cells (Fig. 1) (19, 30), although there is no clear consensus regarding the role of S1/S2 cleavage. The SARS-CoV is similarly sensitive to low endocytic pH, although in this case, there may be an additional fusion requirement mediated following activation of endosomal proteases at sites different from the S1/S2 boundary (48).
Coronaviruses can undergo conformational changes following receptor binding at neutral or slightly alkaline pH (34, 57), and one possibility is that there are multiple triggers to induce fusion during coronavirus entry. Indeed, retroviruses, such as avian leukosis virus (ALV), have been shown to require a combination of receptor priming and low pH for fusion (38). Our fluorescence-dequenching assays for IBV clearly show that the rapid lipid-mixing event in coronavirus fusion is pH dependent, in contrast to the pH-neutral lipid mixing first proposed for ALV using R18-based FdQ assays and virus-cell fusion (12); however, recent work imaging individual fusion events has indicated that low-pH-dependent lipid mixing may occur for ALV (37). It remains possible that SARS-CoV may have additional requirements for fusion activation, e.g., proteolytic cleavage, and that all CoVs require prior receptor interaction for full pH-dependent conformational changes. Currently, our data support the idea that low-pH-dependent conformational changes in IBV can undergo reversion (Fig. 8 and 9). However, it is important to note that in these experiments, reversibility occurs in the absence of receptor, and so it remains to be determined if these changes result in the exposure of the fusion peptide or if they have any direct impact on the fusion event itself. Future work will address these possibilities using our IBV model.
Fluorescence-dequenching studies require purified virus that is stable for purification protocols, and most coronaviruses do not grow to sufficiently high titer in cell culture to allow this type of experiment to be performed. The avian coronavirus IBV that we use here grows to high titer in embryonated chicken eggs and allows purification of sufficient levels of virus for dequenching studies. We believe that IBV is a robust system that can be used to understand fundamental mechanisms of coronavirus biology, allowing sophisticated biochemical and biophysical studies to be performed. The relevance of our model to coronavirus pathogenesis in general is underscored by the similar pathogeneses of IBV and SARS-CoV in their hosts (43, 55); both can cause fatal lung inflammation with secondary bacterial infections, as well as infection of the gastrointestinal tract and kidney (5).
Our data have considerable implications for the development of current and future antiviral therapies for coronavirus infections. Anti-HIV type 1 drugs have recently been approved based on peptides that block its relatively slow receptor-primed fusion reaction at neutral pH (10), and many anti SARS-CoV strategies have involved similar peptidomimetic approaches (2, 26, 31). Although such six-helix-bundle-targeted peptides can have some activity against SARS-CoV fusion, they have not generally been found to be effective anti-SARS inhibitors (52), being functional only at micromolar concentrations, unlike peptides targeting HIV type 1, which are functional at nanomolar concentrations (24). Future antiviral approaches targeting coronavirus fusion may need to take into account a rapid virus-cell fusion reaction with a low-pH-dependent trigger, such as that shown here.
Acknowledgments
We thank Ruth Collins for helpful discussions throughout the course of this work, Brian Crane and David Russell for critical reading of the manuscript, Bhumit Patel for development of Sendai virus assays, Damon Ferguson and Ben Briggs for excellent technical assistance, and all members of the Whittaker laboratory for helpful suggestions.
This work was supported by grant R03 AI060946 from the National Institutes of Health. Work in the authors' laboratory is also supported by a Career Investigator Grant from the American Lung Association (to G.R.W.).
REFERENCES
- 1.Beaudette, F. R., and C. R. Hudson. 1937. Cultivation of the virus of infectious bronchitis. J. Am. Vet. Med. Assoc. 90:51-60. [Google Scholar]
- 2.Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 101:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulay, F., R. W. Doms, I. Wilson, and A. Helenius. 1987. The influenza hemagglutinin precursor as an acid-sensitive probe of the biosynthetic pathway. EMBO J. 6:2643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanagh, D. 2005. Coronaviridae: a review of coronaviruses and toroviruses, p. 1-54. In A. Schmidt, M. H. Wolff, and O. Weber (ed.), Coronaviruses with special emphasis on first insights concerning SARS. Birkhauser Verlag, Basel, Switzerland.
- 6.Cavanagh, D. 1995. The coronavirus surface glycoprotein, p. 73-113. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 7.Cavanagh, D., and P. J. Davis. 1986. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 67:1443-1448. [DOI] [PubMed] [Google Scholar]
- 8.Cavanagh, D., and S. A. Naqi. 2003. Infectious bronchitis, p. 101-119. In S. M. Saif (ed.), Diseases of poultry. Iowa State Press, Ames, Iowa.
- 9.Chambers, P., C. R. Pringle, and A. J. Easton. 1990. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 71:3075-3080. [DOI] [PubMed] [Google Scholar]
- 10.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 11.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell. Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 12.Earp, L. J., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 15.Fabricant, J. 1951. Studies on the diagnosis of Newcastle disease and infectious bronchitis. IV. The use of the serum neutralization test in the diagnosis of infectious bronchitis. Cornell Vet. 61:68-80. [PubMed] [Google Scholar]
- 16.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudin, Y. 2000. Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Subcell. Biochem. 34:379-408. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, J. M., D. Mason, and J. M. White. 1990. Fusion of Rous sarcoma virus with host cells does not require exposure to low pH. J. Virol. 64:5106-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, G. H., B. Delmas, L. Besnardeau, L. K. Vogel, H. Laude, H. Sjostrom, and O. Noren. 1998. The coronavirus transmissible gastroenteritis virus causes infection after receptor-mediated endocytosis and acid-dependent fusion with an intracellular compartment. J. Virol. 72:527-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoekstra, D., and K. Klappe. 1993. Fluorescence assays to monitor fusion of enveloped viruses. Methods Enzymol. 220:261-276. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann, H., and S. Pohlmann. 2004. Cellular entry of the SARS coronavirus. Trends Microbiol. 12:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, K. V. 2003. SARS-associated coronavirus. N. Engl. J. Med. 348:1948-1951. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, K. V., and S. R. Compton. 1995. Coronavirus receptors, p. 55-71. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 24.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 25.Karaca, K., S. Naqi, and J. Gelb, Jr. 1992. Production and characterization of monoclonal antibodies to three infectious bronchitis virus serotypes. Avian Dis. 36:903-915. [PubMed] [Google Scholar]
- 26.Kliger, Y., and E. Y. Levanon. 2003. Cloaked similarity between HIV-1 and SARS-CoV suggests an anti-SARS strategy. BMC Microbiol. 3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korte, T., and A. Herrmann. 1994. pH-dependent binding of the fluorophore bis-ANS to influenza virus reflects the conformational change of hemagglutinin. Eur. Biophys. J. 23:105-113. [DOI] [PubMed] [Google Scholar]
- 28.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication. In D. M. Knipe and P. M. Howely (ed.), Fields virology. Lippincott Wilkins and Williams, Philadelphia, Pa.
- 29.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 277:19727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, D., and D. Cavanagh. 1992. Coronavirus IBV-induced membrane fusion occurs at near-neutral pH. Arch. Virol. 122:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S., G. Xiao, Y. Chen, Y. He, J. Niu, C. R. Escalante, H. Xiong, J. Farmar, A. K. Debnath, P. Tien, and S. Jiang. 2004. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lontok, E., E. Corse, and C. E. Machamer. 2004. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 78:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsh, M., and R. Bron. 1997. SFV infection in CHO cells: cell-type specific restrictions to productive virus entry at the cell surface. J. Cell Sci. 110:95-103. [DOI] [PubMed] [Google Scholar]
- 34.Matsuyama, S., and F. Taguchi. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 76:11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuyama, S., M. Ujike, S. Morikawa, M. Tashiro, and F. Taguchi. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA 102:12543-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMartin, D. 1993. Infectious bronchitis. In J. B. McFerran and M. S. McNulty (ed.), Virus infections of birds. Elsevier, Amsterdam, The Netherlands.
- 37.Melikyan, G. B., R. J. Barnard, L. G. Abrahamyan, W. Mothes, and J. A. Young. 2005. Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proc. Natl. Acad. Sci. USA 102:8728-8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. T. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 39.Ohki, S., T. D. Flanagan, and D. Hoekstra. 1998. Probe transfer with and without membrane fusion in a fluorescence fusion assay. Biochemistry 37:7496-7503. [DOI] [PubMed] [Google Scholar]
- 40.Otsuki, K., K. Noro, H. Yamamoto, and M. Tsubokura. 1979. Studies on avian infectious bronchitis virus (IBV). II. Propagation of IBV in several cultured cells. Arch. Virol. 60:115-122. [DOI] [PubMed] [Google Scholar]
- 41.Payne, H. R., and J. Storz. 1988. Analysis of cell fusion induced by bovine coronavirus infection. Arch. Virol. 103:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedroso de Lima, M. C., J. Ramalho-Santos, M. F. Martins, A. Pato de Carvalho, V. Bairos, and S. Nir. 1992. Kinetic modeling of Sendai virus fusion with PC-12 cells. Effect of pH and temperature on fusion and viral inactivation. Eur. J. Biochem. 205:181-186. [DOI] [PubMed] [Google Scholar]
- 43.Peiris, J. S., Y. Guan, and K. Y. Yuen. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88-S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puri, A., J. Winick, R. J. Lowy, D. Covell, O. Eidelman, A. Walter, and R. Blumenthal. 1988. Activation of vesicular stomatitis virus fusion with cells by pretreatment at low pH. J. Biol. Chem. 263:4749-4753. [PubMed] [Google Scholar]
- 45.Roberts, P. C., T. Kipperman, and R. W. Compans. 1999. Vesicular stomatitis virus G protein acquires pH-independent fusion activity during transport in a polarized endometrial cell line. J. Virol. 73:10447-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosen, C. G., and G. Weber. 1969. Dimer formation from 1-amino-8-naphthalenesulfonate catalyzed by bovine serum albumin. A new fluorescent molecule with exceptional binding properties. Biochemistry 8:3915-3920. [DOI] [PubMed] [Google Scholar]
- 47.Sieczkarski, S. B., and G. R. Whittaker. 2002. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 76:10455-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 102:11876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 101:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 51.Sturman, L. S., C. S. Ricard, and K. V. Holmes. 1990. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37°C correlates with virus aggregation and virus-induced cell fusion. J. Virol. 64:3042-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veiga, S., Y. Yuan, X. Li, N. C. Santos, G. Liu, and M. A. Castanho. 2005. Why are HIV-1 fusion inhibitors not effective against SARS-CoV? Biophysical evaluation of molecular interactions. Biochim. Biophys. Acta 1760:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vennema, H., L. Heijnen, A. Zijderveld, M. C. Horzinek, and W. J. Spaan. 1990. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 64:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weismiller, D. G., L. S. Sturman, M. J. Buchmeier, J. O. Fleming, and K. V. Holmes. 1990. Monoclonal antibodies to the peplomer glycoprotein of coronavirus mouse hepatitis virus identify two subunits and detect a conformational change in the subunit released under mild alkaline conditions. J. Virol. 64:3051-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, M. M., M. Grabe, S. Adams, R. Y. Tsien, H. P. Moore, and T. E. Machen. 2001. Mechanisms of pH regulation in the regulated secretory pathway. J. Biol. Chem. 276:33027-33035. [DOI] [PubMed] [Google Scholar]
- 57.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]