Abstract
Rickettsia felis is maintained transovarially in Ctenocephalides felis fleas in a widespread geographic distribution and is transmitted to humans and animals, including opossums. This rickettsia is phylogenetically a member of the spotted fever group, most closely related to Rickettsia akari and R. australis. An unusual feature of this rickettsia is that the gene for the outer membrane protein A (OmpA) is interrupted by stop codons. To determine if this putatively dying gene is expressed, mRNA was extracted from laboratory-maintained, R. felis—infected cat fleas. Reverse transcriptase—polymerase chain reaction amplification of three segments of the ompA gene indicated that mRNA of ompA is actively transcribed in fleas. The cDNA sequences expressed represented mRNA of the first 1860-basepair segment of Ompa, which includes domains I and II, part of domain III, the region from site 1836 to site 2180, despite the presence of several stop codons, and the open reading frame from site 2788 to site 3837. The detected sequences showed several differences in the amino acid composition when compared with the previously reported sequence.
INTRODUCTION
The family Rickettsiaceae contains many organisms that cause important diseases in humans around the world with a broad range of clinical manifestations. The genus Rickettsia can be divided in two groups according to their antigenic and molecular profiles: the spotted fever group (SFG) and the typhus group.1 Both groups share some antigenic proteins, including a 135-kD outer membrane protein (OmpB) and a 17-kD lipoprotein.2-4
In addition, the SFG has a 190-kD transmembrane protein that contains tandemly repeated sequences flanked by conserved regions.5 The ompA gene has been described in almost all the members of the SFG. The gene length may differ between species depending on the number of repeated sequences, and this genetic variability becomes an excellent tool for molecular characterization among species of Rickettsia or for the identification of different isolates from the same species.6,7 Antigenic variation among Rickettsia species may be related, at least in part, to differences in the number, type, and order of the repeated units of ompA.7
Recombinant constructs containing a fragment of OmpA stimulate protective immunity in mice, and functionally, monoclonal antibodies against OmpA inhibit the adhesion of Rickettsia rickettsii to L-929 cells.8,9 These results support a role of OmpA in cell invasion and reinforce the potential use of this protein for vaccine development.
We reported the DNA sequence of ompA of Rickettsia felis, confirming the taxonomic position of R. felis in the SFG, and described the presence of internal stop codons in the sequence.10 Its situation as a putatively dying gene makes this protein an excellent model for the study of the evolution of the genus and the pathogenic role of the outer membrane protein in rickettsial infection. This study was undertaken to investigate the expression of the sequences of ompA.
The factors involved in gene expression, the regulation of ompA, and the role of this protein in different environments where the rickettsia lives are unknown. Our findings not only support the active transcription of the ompA gene of R. felis in infected vectors, but also demonstrate the potential full transcription of apparently truncated genes in Rickettsia. The methodology strengthens our analysis of rickettsial gene expression for vaccine development and functional assays.
MATERIALS AND METHODS
Infected fleas. Ctenocephalides felis were generously provided by Dr. Lane Foil (Department of Entomology, Louisiana State University Agricultural Center, Baton Rouge, LA). The fleas were maintained frozen at -20°C.10
The fleas were grouped in pools of 50 insects, disinfected with 70% ethanol for 30 minutes, and macerated under sterile conditions in 700 μL of lysis buffer (RNAqueous; Ambion, Austin, TX).
Isolation of RNA from C. felis. Total RNA was extracted from infected fleas using RNAqueous (Ambion), according to the manufacturer's instructions. Total RNA was resuspended in 50 μL of elution buffer provided by Ambion, and DNase I (RNase free) (Ambion) was added to a final concentration of 2 units/μL and incubated at 37°C for 60 minutes. The DNase was inactivated, and RNA extraction was performed with the DNase-free kit (Ambion). One unit per microliter of SUPERase-In (Ambion) was added, and the RNA samples were stored at -80°C.
Reverse transcriptase—polymerase chain reaction (RTPCR). Reverse transcription of the isolated RNA was performed using the Superscript III™ one-step protocol (Super-Script III™ One-Step RT-PCR System; Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The RTPCRs were performed with primers designed for this work (Table 1 and Figure 1). The reactions were performed with 3 μg of total RNA, 0.5 M of each primer, 1× Superscript III buffer, 1 unit/μL of SUPERase-In (Ambion), and RNase-free water to a final volume of 50μL. Reverse transcription was performed at 58°C for 30 minutes with primer set 1, and at 60°C for 30 minutes with the primer sets 2 and 3. The enzyme was heat-inactivated for 2 minutes at 94°C, and PCR conditions were 15 seconds at 94°C, 30 seconds at 58°C, and 2 minutes at 68°C (primer set 1), and 15 seconds at 94°C, 30 seconds at 60°C, and 90 seconds at 68°C (primer sets 2 and 3) for 40 cycles. As a control to ensure that there was no residual DNA or DNA contamination, a PCR was performed on the mRNA template without the inclusion of reverse transcriptase.
TABLE 1.
Primers used to amplify the outer membrane protein A (ompA) gene of Rickettsia felis in this study
| Name | Sequence | Nucleotide position | Reference name |
|---|---|---|---|
| Rf190.1fw | 5′-ATGGCGAATATTTCTCTAAAATTA-3′ | 1-1860 | Primer set 1 |
| Rf190.1800rev | 5′-TTAACTCACCACCACCGTTAGCAAGACCG-3′ | ||
| Rf19.2788fw | 5′-ACATTACCTGCAAACTTTAACTTAACAG-3′ | 2788-3837 | Primer set 2 |
| Rf190.3837rev | 5′-CCGGCAGCTACGGCA-3′ | ||
| Rf190.1836fw | 5′-CTTGCTAACGGTGGTGGTGAGT-3′ | 1836-2180 | Primer set 3 |
| Rf190.2180rev | 5′-CTGACCGTTGGCTAAAATTCCTAC-3′ |
FIGURE 1.
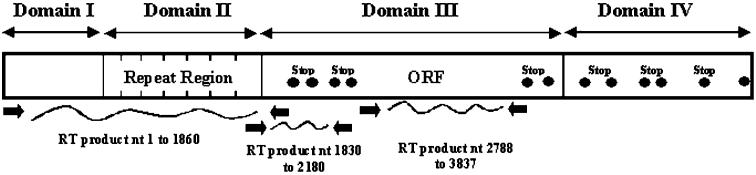
Non-scale schematic representation of the Rickettsia felis outer membrane protein A (ompA) gene. The R. felis ompA sequence (GenBank accession no. AF191026) is represented by the open rectangle. The double head arrows at the top indicate the domains, and the filled circles indicate where the stop codons are present. The filled arrows below indicate the positions of the primers used for the reverse transcriptase— polymerase chain reaction (RT-PCR). ORF = open reading frame; nt = nucleotides.
Sequence of the ompA RT/PCR products. The ompA RT/PCR products were cloned into the Topo TA pCR 2.1 vector (Invitrogen). Selected clones were sequenced three times using a Perkin Elmer (Wellesley, MA) ABI Prism 377 automated sequencer, and the sequences were compared with the ompA genes from other members of the genus Rickettsia in the GenBank database. The sequences were submitted to GenBank (accession numbers AY727036, AY729962, and AY817673).
Sequence data and phylogenetic analysis. The nucleotide and translated sequences from the cDNA products were compared with the sequence of R. felis ompA from colonized fleas reported previously (AF191026) and with the partial sequence of R. felis ompA from field-collected fleas (AJ563398). Sequence comparison was done using the Clustal W program at the European Bioinformatics Institute11 The GenBank accession numbers of the ompA protein gene sequences are R. felis, AF191026;10 R. conorii Malish 7 U01028;5 R. australis AF149108;12 and R. rickettsii M31227.13
Phylogenetic analyses were performed using the maximum-parsimony and distance program of the PAUP 4.1 software.14 Distance matrix analyses were generated with the Kimura two-parameter model for multiple substitutions.15
RESULTS
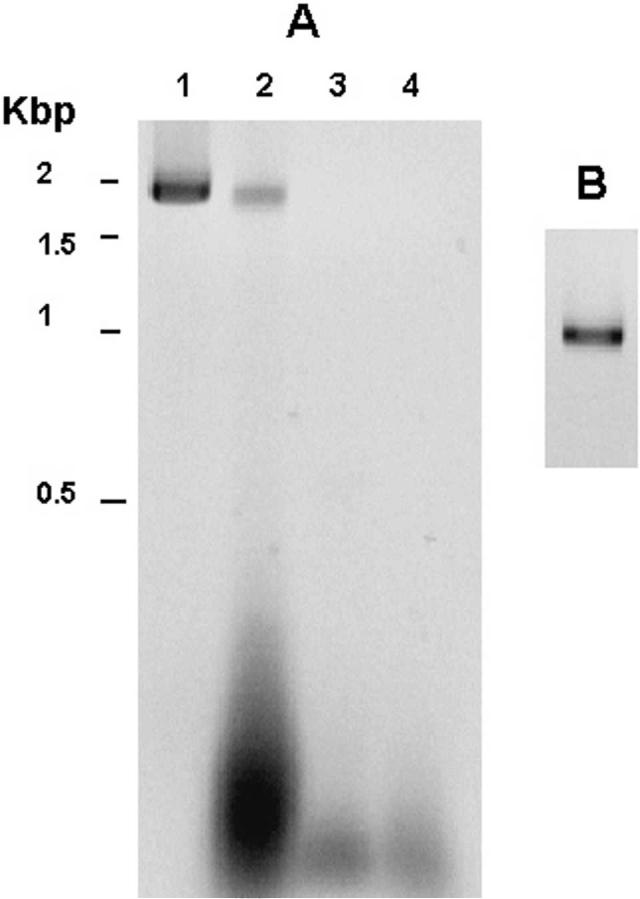
Reverse transcriptase—polymerase chain reaction. To confirm that expression of R. felis ompA was truncated as suggested previously,10 primer sets were designed to detect the expression of the coding region, the region of domain III directly 3 ’ to the stop region that we designated the 3 ’ open reading frame (3 ’-ORF), and for the stop region present between these sequences. The RT-PCR product detected with primer set 1 consisted of the first 1,860-basepair coding region of the ompA sequence that contains domains I, II, and part of domain III according to the terminology of Anderson and others 13 (Figure 2A). The nucleotide sequence of the cDNA fragment was submitted to the European Molecular Biology Laboratory (EMBL) (Heidelberg, Germany) nucleotide database with the accession number AY727036. The sequence showed 99% identity with R. felis and less than 60% homology when compared with the reported ompA sequences of other SFG rickettsiae. The translated cDNA sequence (R. felis 04) showed two differences when compared with the reported R. felis sequence from Mexican field-collected fleas and three differences in amino acid composition in domain II when compared with the published R. felis sequence (Table 2).
FIGURE 2.
Reverse transcriptase—polymerase chain reaction (RTPCR) assay of the outer membrane protein A (ompA) gene of Rickettsia felis. A, RT/PCR assay with total mRNA and ompA primers set 1 (nucleotides 1—1860). Molecular sizes were determined by a 1-kb DNA ladder, Lane 1, genomic PCR; lane 2, RT/PCR product of R. felis RNA; lane 3, water; lane 4 PCR of R. felis RNA without RT (control for potential DNA contamination). Kbp = kilobasepairs. B, RT/PCR with total mRNA using primer set 2 (nucleotides 2788— 3837). Lane 1, RT/PCR product.
TABLE 2.
Amino acid variations in outer membrane protein A (OmpA) between Rickettsia felis, R. felis 04, and R. felis Mexican field-collected source*
| Amino acid position |
|||||||
|---|---|---|---|---|---|---|---|
| 6 | 170 | 242 | 345 | 355 | 957 | 1154 | |
| R. felis† | L | G | P | N | I | G | V |
| R. felis 04† | L | G | S | S | V | D | A |
| R. felis Mexican§ | P | A | - | - | - | - | - |
The positions indicated correspond to the amino acid position from the first methionine (position 1). A filled circle indicates homology, and dashes the unknown aa composition in the R. felis Mexican ompA sequence. - = sequence not available.
R. felis GenBank accession number AF191026.
R. felis 04 GenBank accession number AY727036.
R. felis Mexican GenBank accession number AJ563398.
The RT-PCR product identified with the primer set 2 was a 1,050-basepair segment that corresponded to the region present from nucleotide site 2788 to site 3837 in the ompA sequence, directly 3 ’ downstream to the stop region. The nucleotide sequence of the 3 ’-ORF fragment was submitted to the EMBL nucleotide database with the accession number AY729962 (Figure 2B). The translated sequence showed two differences in amino acid composition when it was compared with the previously reported ompA sequence of R. felis (Table 2).
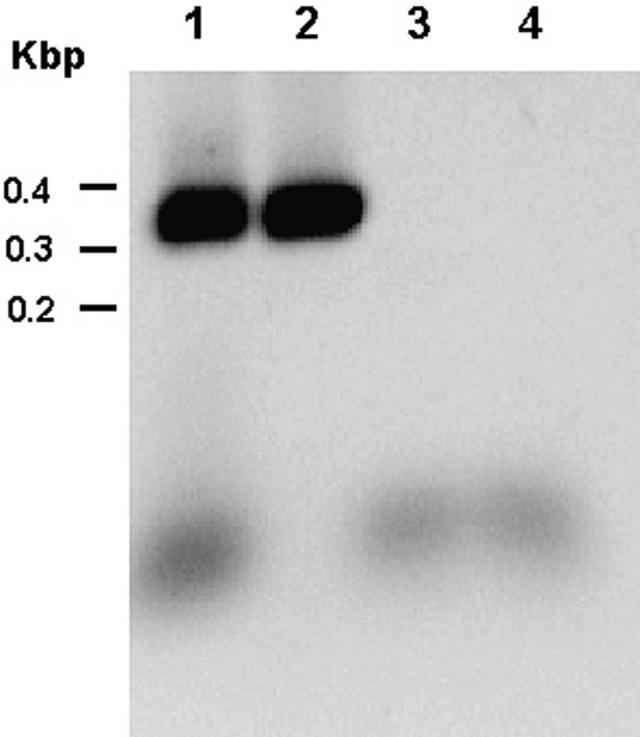
With primer set 3, we detected a 345-nucleotide transcript corresponding to the stop region located between the two sequences detected with the primer sets 1 and 2 (Figure 3). The five-nucleotide differences found in this fragment with respect to the previously reported sequence of R. felis modify the last 30 amino acids in the translated sequence.
FIGURE 3.
Reverse transcriptase—polymerase chain reaction (RTPCR) assay of the stop codon region of the outer membrane protein A (ompA) gene of Rickettsia felis using primer set 3 (nucleotides 1836—2180). Molecular sizes were determined by a 100-basepair DNA ladder. Lane1, RT/PCR product of R. felis RNA; lane 2, genomic PCR; lane 3, water; lane 4, PCR of R. felis RNA without RT (control for DNA contamination). Kbp = kilobasepairs.
Sequence analysis and taxonomy. As we proposed previously,10 we analyzed this transcript according to the protein domains by combining the data from the two sequences outside the repeats. Domain I of each OmpA protein started with the first methionine (residue 1) and ended at the beginning of the repeat region. The 3 ’-ORF region started with the first amino acid after the last stop codon (amino acid position 930 of the translated sequence) and ended at the last amino acid present in this ORF just before the next stop codon region (amino acid position 1288). The combined analysis of the domain I and the 3 ’-ORF in domain III of OmpA represented 968 amino acid residues.
The phylogenetic distances found for both R. felis sequences in this analysis were similar when compared with other SFG rickettsiae (Table 3), but were not identical. Both cDNA and genomic sequences showed slight differences that represent nearly 0.2% of the sequences analyzed.
TABLE 3.
Amino acid pair distance of outer membrane protein A of Rickettsia species
DISCUSSION
Among the immunodominant surface proteins present in SFG rickettsiae, OmpA is considered as one of the most important. Because its presence discriminates between the two groups of the genus Rickettsia, it is used for diagnosis and strain and species characterization, and it is functionally involved in the pathogenesis of SFG rickettsial diseases.9
Although OmpA has been identified in almost all the members of the SFG and has been well characterized at the genomic level, active gene expression had only been demonstrated by indirect approaches such as immunofluorescence and immunoblotting. Previous studies of ompA and expression of other rickettsial proteins had been performed using plasmid vectors3,16
The 1,860-basepair sequence of the ompA cDNA contains domains I, II and part of domain III, and the 1,050-basepair transcript contains the domain III ORF present 3 ’ downstream of the stop codon region (Figure 1). Molecular heterogeneity is denoted by the difference of five amino acids and is similar to the intrastrain variation described in the amino acid composition of the ompB protein of different R. prowazekii strains.17 The differences of the ompA sequences of R. felis obtained from colonized fleas and the 170 amino acid partial sequence of R. felis ompA from field-collected fleas suggest that molecular heterogeneity also occurs in nature and likely represents the existence of different strains of R. felis. Although the occurrence of mutations has not been explored in the genome of any rickettsia maintained in colonized fleas, drastic changes were described in the ompA gene repeat domain of R. rickettsii that apparently developed during prolonged passages in cell culture.18 However, we cannot eliminate the possibility that the changes are the consequence of natural selective pressure during prolonged transovarial maintenance in a flea colony rather than an ecologic cycle also involving a mammalian host. This observation emphasizes the importance of further analysis of wild-type strains of R. felis obtained from fleas and animals infected in nature to determine the natural diversity of the ompA gene and whether ompA truncation is associated with long-term laboratory maintenance of fleas.
The amino acid changes in OmpA of R. felis were serine for proline, serine for asparagine, valine for isoleucine, aspartic acid for glycine, and alanine for valine with respect to the ompA sequence reported previously, and a proline for leucine and an alanine for glycine with respect to the reported 170 amino acid sequence of the Mexican wild-type R. felis ompA sequence (Table 2). These changes may have implications for immune recognition and/or function because two changes alter two amino acids to those of different groups (serine for proline, and aspartic acid for glycine). The slight differences in structure and biochemical properties among the others could also be important for antigenic recognition as reported in other organisms.19-21 Moreover, the sequences of R. felis show huge differences when compared with other rickettsial ompA sequences, and these variations could be used for diagnostic purposes using the protein fragments for detection of antibodies to R. felis in human patients and in animal reservoirs.
The stop region RT fragment contained five nucleotide differences when compared with the reported ompA gene of R. felis, including two deletions, one insertion, and two transversions. The indels and substitutions resulted in a change in the amino acid sequence of the last 30 amino acids in the sequence.
The genome of the genus Rickettsia contains several pseudogenes and ORFs with apparently noncoding function.22 The presence of these sequences suggests an active genome degradation process as a result of an adaptation of the different species to the diverse environments where they live, and could most likely be the result of speciation.22,23 As in other reported pseudogene sequences in many Rickettsia species, the ompA gene of R. felis contains several stop codons that interrupt their reading frame. The accumulation of these stop codons could be the result of the ongoing degradation process to become a pseudogene.
However, at this moment of the evolution of R. felis, the organization of the ompA gene resembles more a split gene than a pseudogene. It had been suggested that some of the truncated genes reported in the genus Rickettsia could retain some functional activity, such as the genes that encode the SpoT proteins of R. prowazekii, which represent a truncated version of the SpoT protein of Escherichia coli.22,24,25
Different transcription patterns with diverse complexity of several split genes had been described previously.24 These results suggest different transcription mechanisms for these genes, but also could reflect the presence of transcripts in different stages of processing. Our results support the concept of a full transcription event in the split genes despite the presence of stop codons or the noncoding region present between the ORFs.
The presence of the ompA transcript raises the possibility of its expression regardless of the condition of being a truncated gene, similar to that of R. peacockii, which has been found in nature only in ticks. A short sequence (600 basepairs) of the truncated ompA gene is detected.26 The putative OmpA protein of R. felis does not have the beta peptide for its transport to the outer membrane, but still has the signal peptide for its insertion into the inner membrane. The absence of the autotransporter segment makes this protein lack the properties needed to function for its own export to the outer membrane, and we are unsure if the remaining inner membrane segment is functional. Moreover, the changes do not affect the transcription terminator region of the gene, and it is also possible that the full sequence is transcribed. This observation is based on the RT-PCR amplification of the stop codon region and the domain III ORF adjacent to the 3 ’ end.
Two predominant heat resistant proteins with molecular masses of 60 and 30 kD have been reported in R. felis.4,27 The molecular weights of these proteins are similar to the masses predicted for the transcript of 1,860 basepairs and the transcript of the 3 ’ ORF reported in this work (60.5 and 36 kD, respectively). Confirmation of this possibility would require further experimental evidence; the presence of both OmpA segments in R. felis would support the hypothesis of the functionality of some split genes by independent units or working together as subunits of one protein.
The gene degradation of ompA in R. felis could have several explanations. As discussed earlier in this report, the presence of this phenomenon could be associated with long-term laboratory maintenance in fleas where the protein might not require the function needed in the mammalian host. The degradation could also have been produced by evolutionary adaptation to their environment and/or could be the result of evolutionary natural genetic heterogeneity among strains.
The phylogenetic relationship of this R. felis variant to other SFG Rickettsia species is the same as reported for R. felis previously (Table 3)10; the differences between the R. felis sequences could be the result of strain variation. The sequence alignment of the R. felis ompA with the partial sequence of the R. akari ompA available in the GenBank (AY727036) was not useful in determining potentially shared segments between the sequences of these two SFG rickettsiae not transmitted by ticks.
In this work, we report for the first time the full-length transcript of a putative dying gene detected by RT/PCR in infected fleas. This study was performed with an adapted methodology that could be considered a contribution to the study of gene expression in members of the genus Rickettsia and related intracellular bacteria, especially when axenic culture is not available.
The expression and isolation of OmpA in vitro is essential to determine its function in the pathogenesis of rickettsial diseases and to develop better methods for diagnosis and epidemiologic surveillance. It is also a potential target for new strategies for developing drugs and vaccines for the control of human illnesses caused by rickettsiae.
Acknowledgments
This research was supported by grants from the Fogarty International Center of the National Institutes of Health (D43 TW00903) and CONACyT (34436-M).
REFERENCES
- 1.Anacker RL, Mann RE, Gonzales C. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J Clin Microbiol. 1987;25:167–171. doi: 10.1128/jcm.25.1.167-171.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan M-Y, Yu X-J, Walker DH. Antigenic analysis of Chinese strains of spotted fever group rickettsiae by protein immunoblotting. Am J Trop Med Hyg. 1988;39:497–501. doi: 10.4269/ajtmh.1988.39.497. [DOI] [PubMed] [Google Scholar]
- 3.Walker DH, Feng HM, Saada JI, Crocquet-Valdes P, Radulovic S, Popov VL, Manor E. Comparative antigenic analysis of spotted fever group rickettsiae from Israel and other closely related organisms. Am J Trop Med Hyg. 1995;52:569–576. doi: 10.4269/ajtmh.1995.52.569. [DOI] [PubMed] [Google Scholar]
- 4.Fang R, Raoult D. Antigenic classification of Rickettsia felis by using monoclonal and polyclonal antibodies. Clin Diag Lab Immunol. 2003;10:221–228. doi: 10.1128/CDLI.10.2.221-228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crocquet-Valdes PA, Weiss K, Walter DH. Sequence analysis of the 190-kDa antigen-encoding gene of Rickettsia conorii (Malish 7 strain) Gene. 1994;140:115–119. doi: 10.1016/0378-1119(94)90740-4. [DOI] [PubMed] [Google Scholar]
- 6.Fournier P-E, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48:839–849. doi: 10.1099/00207713-48-3-839. [DOI] [PubMed] [Google Scholar]
- 7.Gilmore RD., Jr Comparison of the rompA gene repeat regions of Rickettsiae reveals species-specific arrangements of individual repeating units. Gene. 1993;125:97–102. doi: 10.1016/0378-1119(93)90752-o. [DOI] [PubMed] [Google Scholar]
- 8.Crocquet-Valdes PA, Diaz-Montero CM, Feng HW, Li H, Barrett ADT, Walker DH. Immunization with a portion of rickettsial outer membrane protein A stimulates protective immunity against spotted fever group rickettsiosis. Vaccine. 2002;20:979–988. doi: 10.1016/s0264-410x(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Walker DH. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog. 1998;24:289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- 10.Bouyer DH, Stenos J, Crocquet-Valdes P, Moron CG, Popov VL, Zavala-Velazquez JE, Foil LD, Stothard DR, Azad AF, Walker DH. Rickettsia felis: molecular characterization of a new member of the spotted fever group. Int J Syst Evol Microbiol. 2001;51:339–347. doi: 10.1099/00207713-51-2-339. [DOI] [PubMed] [Google Scholar]
- 11.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenos J, Walker DH. The rickettsial outer-membrane protein A and B genes of Rickettsia australis, the most divergent rickettsia of the spotted fever group. Int J Syst Evol Microbiol. 2000;50:1775–1779. doi: 10.1099/00207713-50-5-1775. [DOI] [PubMed] [Google Scholar]
- 13.Anderson BE, McDonald GA, Jones DC, Regnery RL. A protective protein antigen of Rickettsia rickettsii has tandemly repeated near-identical sequences. Infect Immun. 1990;58:2760–2769. doi: 10.1128/iai.58.9.2760-2769.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swofford DL. Version 4. Sinauer Associates; Sunderland, MA: 1998. PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods) [Google Scholar]
- 15.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Policastro PF, Hackstadt T. Differential activity of Rickettsia rickettsii ompA and ompB promoter regions in a heterologous reporter gene system. Microbiology. 1994;140:2941–2949. doi: 10.1099/13500872-140-11-2941. [DOI] [PubMed] [Google Scholar]
- 17.Moron CG, Bouyer DH, Yu X-J, Foil LD, Crocquet-Valdes P, Walker DH. Phylogenetic analysis of the rompB genes of Rickettsia felis and Rickettsia prowazekii European human and North American flying-squirrel strains. Am J Trop Med Hyg. 2000;62:598–603. doi: 10.4269/ajtmh.2000.62.598. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto M, Tange Y, Okada T, Inoue Y, Horiuchi T, Kobayashi Y, Fujita S. Deletion in the 190 kDa antigen gene repeat region of Rickettsia rickettsii. Microb Pathog. 1996;20:57–62. doi: 10.1006/mpat.1996.0005. [DOI] [PubMed] [Google Scholar]
- 19.Zeder-Lutz G, Altschuh D, Denery-Papini S, Briand JP, Tribbick G, van Regenmortel MH. Epitope analysis using kinetic measurements of antibody binding to synthetic peptides presenting single amino acid substitutions. J Mol Recognit. 1993;6:71–79. doi: 10.1002/jmr.300060205. [DOI] [PubMed] [Google Scholar]
- 20.Gomara MJ, Ercilla G, Alsina MA, Haro I. Assessment of synthetic peptides for hepatitis A diagnosis using biosensor technology. J Immunol Methods. 2000;246:13–24. doi: 10.1016/s0022-1759(00)00295-7. [DOI] [PubMed] [Google Scholar]
- 21.Mahler E, Sepulveda P, Jeannequin O, Liegeard P, Gounon P, Wallukat G, Eftekhari P, Levin MJ, Hoebeke J, Hontebeyrie M. A monoclonal antibody against the immunodominant epitope of the ribosomal P2beta protein of Trypanosoma cruzi interacts with the human beta 1-adrenergic receptor. Eur J Immunol. 2001;31:2210–2216. doi: 10.1002/1521-4141(200107)31:7<2210::aid-immu2210>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 22.Andersson JO, Andersson SGE. Pseudogenes, junk, and the dynamics of rickettsia genomes. Mol Biol Evol. 2001;18:829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- 23.Andersson JO, Andersson SGE. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- 24.Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, Roux V, Cossart P, Weissenbach J, Claverie JM, Raoult D. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 25.McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, Fox GE, Jiang H, NcNeill TZ, Muzny D, Jacob LS, Hawes AC, Sodergren E, Gill R, Hume J, Morgan M, Fan G, Amin AG, Gibbs RA, Hong CH, Yu XJ, Walker DH, Weinstock GM. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldridge GD, Burkhardt NY, Simser JA, Kurtti TJ, Munderloh UG. Sequence and expression analysis of the ompA gene of Rickettsia peacockii, an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni. Appl Environ Microbiol. 2004;70:6628–6636. doi: 10.1128/AEM.70.11.6628-6636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raoult D, La Scola B, Enea M, Fournier PE, Roux V, Fenollar F, Galvao MAM, de Lamballerie X. A flea-associated rickettsia pathogenic for humans. Emerg Infect Dis. 2003;7:73–81. doi: 10.3201/eid0701.010112. [DOI] [PMC free article] [PubMed] [Google Scholar]