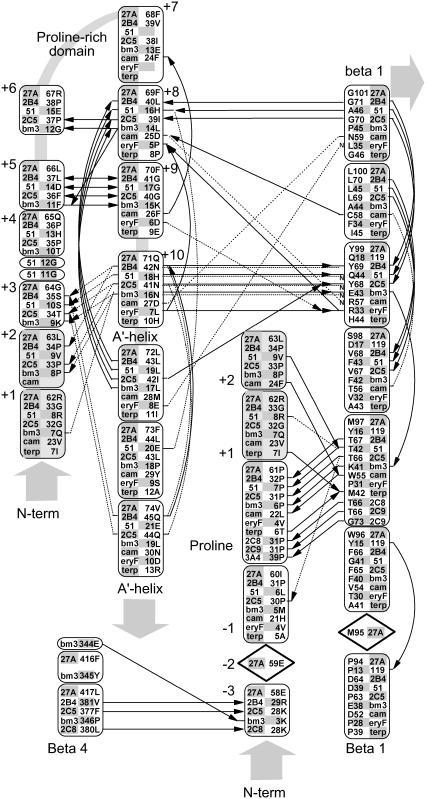
FIGURE 2.
Hydrogen bonding patterns in the N-terminal domain of selected CYP crystallographic templates. Although the structure of the N-terminal domain is highly variable, a conserved hydrogen bond is seen in most templates between a conserved proline amide carbonyl and the amide nitrogen of an aligned β-1 residue. This proline is part of a PGP-motif or larger proline-rich domain found in many P450s but seen as Pro-61, Arg-62, and Leu-63 in Cyp27A1. This proline provides a useful landmark for alignment and modeling and helps position a highly structured hairpin near the β-1 sheet. Relative to this proline landmark, a hydrogen bond is seen from the −3proline amide carbonyl to the β-4 sheet in 4/11 templates. Other hydrogen bonds stabilize this hairpin through +5proline to +9proline double backbone interaction and a +8proline amide carbonyl to +11proline amide nitrogen. Additional hydrogen bonds connect the +10proline backbone and side chain (usually asparagine) to the +2proline and +3proline backbone and the amide nitrogen at a conserved position in the β-1 sheet position. The aligned +1proline and +2proline positions were repeated in the figure for clarity. Notes: i), hydrogen bonding patterns in CYP2C8, CYP2C9, and CYP3A4 are similar to CYP2B4 and CYP2C5; and ii), solid lines indicate backbone hydrogen bonds between amide nitrogen and amide carbonyl (indicated by an arrowhead). Double arrows indicate the presence of two backbone hydrogen bonds between the indicated residues. Dashed lines indicate hydrogen bonds involving side chains, and those interacting with amide nitrogen are marked with a small font “N”. Diamond placeholders indicate positions of aligned residues not involved in structural hydrogen bonding interactions.