Abstract
The genome of mitochondria encodes a small number of very hydrophobic polypeptides that are inserted into the inner membrane in a cotranslational reaction. The molecular process by which mitochondrial ribosomes are recruited to the membrane is poorly understood. Here, we show that the inner membrane protein Mba1 binds to the large subunit of mitochondrial ribosomes. It thereby cooperates with the C-terminal ribosome-binding domain of Oxa1, which is a central component of the insertion machinery of the inner membrane. In the absence of both Mba1 and the C-terminus of Oxa1, mitochondrial translation products fail to be properly inserted into the inner membrane and serve as substrates of the matrix chaperone Hsp70. We propose that Mba1 functions as a ribosome receptor that cooperates with Oxa1 in the positioning of the ribosome exit site to the insertion machinery of the inner membrane.
Keywords: Mba1, membrane insertion, mitochondria, Oxa1, protein synthesis
Introduction
The synthesis of hydrophobic membrane proteins in a hydrophilic environment is a delicate problem as it bears the high risk of the formation of unproductive protein aggregates. Cells typically tackle this task by strictly coupling translation to membrane integration. In the cytosol of eukaryotic and prokaryotic cells, signal recognition particles recognize ribosomes, which synthesize hydrophobic polypeptides and target them to membrane-embedded translocation complexes (Gilmore and Blobel, 1983). These translocases contact the ribosomes in proximity to their polypeptide exit tunnels (Beckmann et al, 1997, 2001; Ménétret et al, 2000). Thereby, hydrophobic translation products are inserted into the hydrophobic environment of the membranes right after they emerge from the ribosome.
While signal recognition particles ubiquitously exist in the cytosol of bacteria and eukaryotes as well as in chloroplasts, they appear to be absent from mitochondria (Glick and von Heijne, 1996). This is particularly surprising as mitochondrial genomes encode almost exclusively highly hydrophobic membrane proteins (Borst and Grivell, 1978) and, thus, the mitochondrial protein synthesis apparatus should be specialized for the production of hydrophobic polypeptides. In the yeast Saccharomyces cerevisiae, eight proteins are encoded by the mitochondrial genome, seven of which are highly hydrophobic membrane proteins: cytochrome b of the bc1-complex, Cox1, Cox2 and Cox3 of the cytochrome oxidase and Atp6, Atp8 and Atp9 of the F1F0-ATPase (Tzagoloff and Myers, 1986; Grivell et al, 1999). The highly hydrophobic nature of these polypeptides apparently prevented the transfer of their genes to the nucleus (Claros et al, 1995) and forced eukaryotic cells to maintain a separate genome in mitochondria.
The insertion of mitochondrial translation products into the inner membrane of mitochondria depends on the conserved inner membrane protein Oxa1. The Oxa1 protein of the budding yeast S. cerevisiae was the founding member of a large protein family, called the Alb3/Oxa1/YidC family, with members in chloroplasts, mitochondria and bacteria (Bauer et al, 1994; Bonnefoy et al, 1994). All homologs share a hydrophobic core domain, which facilitates the insertion and/or folding of membrane proteins (Stuart, 2002; Herrmann and Neupert, 2003; Kuhn et al, 2003). The mitochondrial Oxa1, in contrast to its bacterial homologs, exposes a C-terminal α-helical stretch of about 100 residues into the matrix that functions as ribosome-binding domain (Jia et al, 2003; Szyrach et al, 2003; Preuss et al, 2005). This region contacts the ribosome close to the protein Mrpl20/Mrp20, the homolog of the bacterial L23 protein, which is located next to the polypeptide exit site of the mitochondrial ribosome (Jia et al, 2003). The binding of Oxa1 to the ribosome thereby ensures intimate proximity of the emerging nascent chain to the membrane-embedded core domain of Oxa1.
Oxa1 is supported in its function in protein insertion by an additional mitochondrial protein, Mba1, the function of which is poorly understood. Mba1 is associated with the matrix face of the inner membrane (Rep and Grivell, 1996; Preuss et al, 2001) and was initially found in a genetic screen as multicopy suppressor of mutants of mitochondrial proteases (Rep et al, 1996). Like Oxa1, which was recovered in the same screen, Mba1 is in direct proximity to nascent translation products in mitochondria (Preuss et al, 2001). The deletion of Mba1 leads to insertion defects of mitochondrially encoded proteins. Deletions of both Oxa1 and Mba1 show severe synthetic growth defects, suggesting that both proteins overlap in some aspect of their function.
Here, we show that Mba1 binds to mitochondrial ribosomes. This interaction does neither require the presence of Oxa1 nor that of nascent chains on the ribosomes. In its ribosome-binding function, Mba1 cooperates with the C-terminal domain of Oxa1. Our data suggest that mitochondrial ribosomes are associated with the inner membrane by multivalent interactions to several tethering factors, which, in a concerted manner, coordinate cotranslational membrane insertion in mitochondria.
Results
Mba1 binds to mitochondrial ribosomes
Mba1 was reported to be in close vicinity to nascent chains on mitochondrial ribosomes (Preuss et al, 2001). This inspired us to test whether Mba1 undergoes a physical interaction with ribosomes. In order to isolate ribosomes, mitochondria were prepared from wild-type yeast cells and lysed in 1% Triton X-100. The extract was cleared by centrifugation and layered on top of a linear 0.3–1 M sucrose gradient. After centrifugation, the gradient was fractionated and the protein content of the fractions was analyzed by Western blotting (Figure 1A). Ribosomal proteins migrated deep into the gradient and proteins of the small (Mrps51/Mrp51) (Green-Willms et al, 1998) and large (Mrpl20) subunit were found in different regions of the gradient. Like Oxa1, Mba1 comigrated with Mrpl20, a protein of the large subunit of the mitochondrial ribosome. Treatment of the mitochondrial extracts with high concentrations of RNase led to the disintegration of the ribosomes and prevented the migration of Mba1 and Oxa1 into the gradient (Figure 1B). Even low concentrations of RNase, which only degrade the RNA that is exposed to the ribosomal surface, released Mba1 and Oxa1 largely from ribosomes (Figure 1C). In summary, we conclude that Mba1, like Oxa1, is physically associated with mitochondrial ribosomes.
Figure 1.
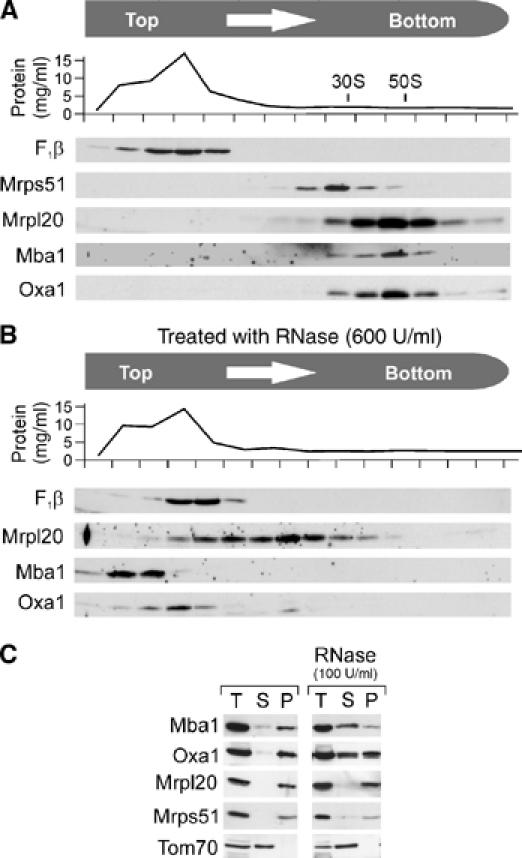
Mba1 cofractionates with mitochondrial ribosomes on sucrose gradients. (A) Wild-type mitochondria (1 mg) were lysed in Triton buffer and the extract was subfractionated by velocity centrifugation on a continuous sucrose gradient. After fractionation of the gradient, the protein concentration in each fraction was determined. The distribution of the following proteins in the fractions was assessed by Western blotting: F1β, the β-subunit of the ATPase; Mrps51 and Mrpl20, proteins of the small and large subunit of the mitochondrial ribosome, respectively; Mba1 and Oxa1. (B) Like A, with the exception that the mitochondrial extract was treated with 600 U/ml of bovine pancreatic RNase A for 30 min to disintegrate mitochondrial ribosomes. (C) Mitochondrial extracts were mock treated or incubated with 100 U/ml of RNase A to remove surface-exposed ribosomal RNA. Remaining ribosomal particles (P) were separated from the supernatant (S) by centrifugation through a high sucrose cushion. Unfractionated samples (T, total) are shown for control.
The binding of Mba1 to ribosomes requires neither nascent chains nor Oxa1
Since Mba1 and Oxa1 can both be efficiently crosslinked to mitochondrial nascent chains (Hell et al, 1997; Preuss et al, 2001), it is conceivable that Mba1 interacts with ribosomes via Oxa1 or via the newly synthesized polypeptides. In order to test this, the cofractionation of Mba1 with ribosomes was assessed in an oxa1 deletion mutant (Δoxa1). As shown in Figure 2A, Mba1 was still associated to ribosomes in the absence of Oxa1. Moreover, Mba1 even remained bound to ribosomes following treatment with puromycin (Figure 2A, right panel). As shown in Figure 2B, the treatment with puromycin released radiolabeled nascent chains from mitochondrial ribosomes, which then were rapidly degraded (Pajic et al, 1994; Arlt et al, 1996). In contrast, without the addition of puromycin nascent chains remained ribosome-associated upon lysis both in the presence or absence of Mg2+ and only were released upon incubation with high concentrations of salt. In summary, Mba1 binds mitochondrial ribosomes independently of Oxa1 and ribosome-associated nascent chains.
Figure 2.
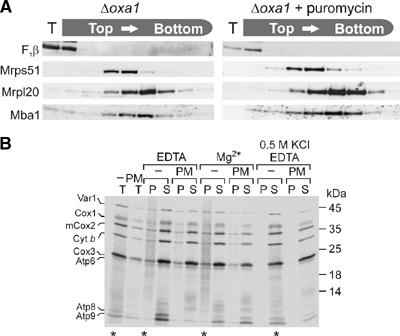
Ribosome binding of Mba1 is independent of Oxa1 and nascent chains. (A) Mitochondria (234 μg) isolated from Δoxa1 cells were incubated in translation buffer for 15 min. The sample was split and to one-half 2 mM puromycin was added. Following incubation for 5 min, the mitochondria were lysed and fractionated as described in Figure 1. The distribution of proteins was assessed by Western blotting. (B) Translation products were radiolabeled in isolated wild-type mitochondria (400 μg) for 15 min. The sample was split and one-half treated with puromycin (PM) as in (A). The reactions were divided into four aliquots. Mitochondria were directly applied to the gel (T, total) or lysed in 1% Triton X-100, 20 mM Tris, pH 7.4, containing 5 mM EDTA, 50 mM KCl (EDTA) or 5 mM Mg2+, 50 mM KCl (Mg2+) or 5 mM EDTA, 0.5 M KCl (0.5 M KCl EDTA). The extracts were cleared by centrifugation and applied onto a 1.2 M sucrose cushion. A ribosomal pellet fraction (P) was separated from residual proteins in the supernatant (S) by centrifugation for 60 min at 200 000 g. Proteins were resolved by SDS–PAGE and visualized by autoradiography. Fractions containing nascent chains, which appear as a smear due to their heterogenic size, are indicated by asterisks.
Recombinant Mba1 binds mitochondrial ribosomes in vitro
To assess more directly the binding of Mba1 to mitochondrial ribosomes by affinity chromatography, Mba1 was expressed in Escherichia coli as a fusion protein to maltose-binding protein (MBP). This recombinant MBP-Mba1 fusion protein and unfused MBP as control were immobilized on amylose beads. Mitochondrial translation products were radiolabeled in isolated mitochondria. The mitochondria were lysed and the extract was incubated with the immobilized MBP fusion proteins. Following extensive washing of the resin, proteins bound to the fusion protein were eluted with sample buffer, subjected to SDS–PAGE and analyzed by autoradiography and Western blotting (Figure 3A). Ribosomal proteins of the large (Mrpl36, a homolog of the bacterial L31 protein) and the small ribosomal subunit (Mrps51) as well as the radiolabeled nascent chains were specifically recovered with the MBP-Mba1 protein but not with the MBP control (Figure 3A, lanes 2 and 3). We conclude that Mba1 has the ability to bind actively translating mitochondrial ribosomes in vitro. Unrelated mitochondrial proteins like aconitase and Tom70 were not bound. Interestingly, also Oxa1 was pulled down specifically with the MBP-Mba1 fusion protein, although it had been shown before that Mba1 and Oxa1 form physically distinct complexes in mitochondria (Preuss et al, 2001).
Figure 3.
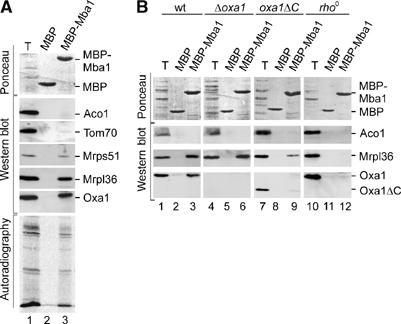
Mba1 binds mitochondrial ribosomes in vitro. (A) Recombinant MBP and MBP-Mba1 were purified and immobilized on an amylose resin. Mitochondrial translation products were radiolabeled in isolated mitochondria. Mitochondria were solubilized. The resulting extract was clarified by centrifugation and subjected to the immobilized MBP fusion proteins. Following extensive washing, bound proteins were eluted with sample buffer, resolved by SDS–PAGE and transferred to nitrocellulose. Transferred proteins were analyzed by staining with Ponceau S (upper panel) or by Western blotting with antibodies against proteins of the large (Mrpl36) and small (Mrps51) ribosomal subunit, Oxa1, aconitase (Aco1) and Tom70. The lower panel shows the radioactive translation products that were detected by autoradiography. Total lanes (T) show 10% of the mitochondrial extract that was applied to the resin. (B) Translation products were radiolabeled in mitochondria isolated from wild type (wt), a Δoxa1 mutant, a mutant containing a C-terminally truncated variant of Oxa1 (oxa1ΔC) and a rho0 strain. The binding of ribosomes and Oxa1 to the immobilized MBP-Mba1 fusion protein was assessed as in (A).
To characterize in more detail the molecular interactions between Mba1, ribosomes and Oxa1, we applied mitochondrial extracts of different yeast strains to the bait protein. First, we tested the relevance of Oxa1 for the interaction of ribosomes with the immobilized Mba1. Therefore, we generated extracts that lacked either Oxa1 entirely or the C-terminal 71 amino acid residues (Figure 3B, lanes 4–9). In both cases, the ribosomes still bound to Mba1. However, the C-terminally truncated version of Oxa1 was not recovered with the bound fractions. Thus, the C-terminal ribosome-binding domain of Oxa1 was apparently critical for its interaction with Mba1, suggesting that Oxa1 did not directly bind to Mba1 but via its interaction to the ribosome. This was further supported by experiments with extracts of mitochondria lacking mitochondrial DNA (rho0). In these extracts, which due to the absence of rRNA do not contain assembled ribosomes, the binding of Oxa1 was prevented (Figure 3B, lanes 10–12). Hence, the presence of ribosomes appears necessary for the interaction of Mba1 with Oxa1. In the rho0 extracts, the ribosomal protein Mrpl36 was not recovered with Mba1, excluding its unspecific binding to the MBP-Mba1 bait protein. We conclude that Mba1 has the ability to bind mitochondrial ribosomes and, indirectly via the ribosome, to the Oxa1 translocase. Thus, Mba1 and Oxa1 appear to serve as membrane anchors for the mitochondrial ribosome, which can bind the ribosome simultaneously and independently of each other.
Mba1 and Oxa1 mutants show synthetic insertion defects
The observed binding of Mba1 to mitochondrial ribosomes suggests that Mba1 supports Oxa1 in its function to tether translating ribosomes to the inner membrane. Indeed, the simultaneous deletion of both Mba1 and the C-terminal 71 residues of Oxa1 led to severe synthetic growth defects on the nonfermentable carbon source glycerol, whereas the single deletions showed only very mild defects (Figure 4A). No growth defects were observed for these mutants on the fermentable carbon source glucose, for which respiratory activity is not critical.
Figure 4.
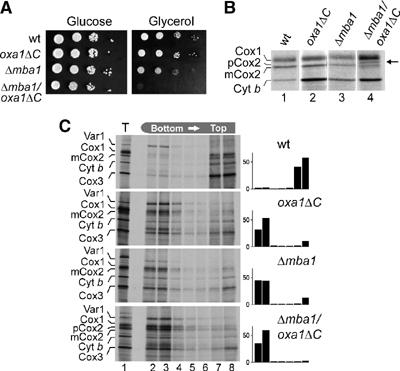
Deletion of Mba1 and the ribosome-binding domain of Oxa1 causes synergistic growth and insertion defects. (A) The strains indicated were grown on YPD medium to log phase. Serial 10-fold dilutions of the cultures were spotted on YP plates containing 2% glucose or 2% glycerol and plates were incubated at 30°C for 2 or 3 days, respectively. (B) The deletion of both Mba1 and the ribosome-binding domain of Oxa1 leads to the accumulation of unprocessed Cox2. Translation products were radiolabeled in mitochondria isolated from the strains indicated for 10 min at 25°C, separated by SDS–PAGE and visualized by autoradiography. (C) The deletion of Mba1 or of the ribosome-binding domain of Oxa1 causes a general insertion defect of mitochondrial translation products. Translation products were radiolabeled in mitochondria isolated from the strains indicated for 30 min at 30°C. The mitochondria were reisolated, washed and treated with 0.1 M Na2CO3 and 4.5 M urea. The suspension was adjusted to 1.6 M sucrose and layers of 1.4 and 0.25 M sucrose were placed on top of it. The gradient was centrifuged and fractionated. Proteins in the fractions were precipitated with 12% trichloroacetic acid and analyzed by SDS–PAGE and autoradiography. The radioactive signals of the translation products were quantified in each fraction and are depicted on the right.
To assess the insertion of mitochondrial translation products in these strains, we tested the electrophoretic mobility of newly synthesized Cox2. Subunit 2 of cytochrome oxidase is synthesized in the matrix with a presequence, which is removed by the Imp1 protease, a homolog of the bacterial leader peptidase, upon translocation to the intermembrane space. The accumulation of the precursor form of Cox2 is therefore indicative of defects in the insertion process of mitochondrial translation products. As shown in Figure 4B, the deletion of either Mba1 or of the C-terminus of Oxa1 did not compromise the processing of Cox2. In contrast, considerable amounts of Cox2 precursor accumulated in the double mutant (Figure 4B, lane 4, arrowhead). This indicates that the insertion of Cox2 is hampered if both Mba1 and the C-terminus of Oxa1 are absent.
Next, we performed fractionation experiments to determine the insertion defect of the Δmba1/oxa1ΔC double mutant in more detail. Translation products were radiolabeled in isolated mitochondria and proteins that are not tightly associated with membranes were extracted by incubation with carbonate and urea. Extractable (soluble and membrane-associated) and nonextractable (membrane-embedded) proteins were separated by flotation gradient centrifugation (Figure 4C). In wild-type mitochondria, the ribosomal subunit Var1 is the only protein detected in the soluble fractions at the bottom of the gradient; all other translation products and the nascent chains fractionated with the membranes on the top of the gradients (Figure 4C, upper panel, lanes 7–8). With mutants bearing individual deletions of Mba1 or the C-terminus of Oxa1, significant amounts of the translation products were shifted to the fractions with extractable proteins (Figure 4C, second and third panel, lanes 2–4). This shows defective membrane integration in both mutants. The failure to insert membrane proteins properly was more pronounced when both Mba1 and the ribosome-binding domain of Oxa1 were absent (Figure 4C, lower panel, lanes 7–8). This suggests that Mba1 and the C-terminus of Oxa1 cooperate in the cotranslational insertion of newly synthesized translation products in mitochondria. Presumably, both factors play a pivotal role in the positioning of the polypeptide exit tunnel to the sites of protein insertion in the inner membrane.
In the absence of Mba1 and the C-terminus of Oxa1, hydrophobic translation products are associated with Hsp70
The mitochondrial Hsp70 (mtHsp70) protein is part of a chaperone system, which specifically interacts with unfolded, non-native polypeptides in the matrix space (Hartl and Hayer-Hartl, 2002). The hydrophobic membrane proteins synthesized on mitochondrial ribosomes are normally not found in contact with mtHsp70 (Herrmann et al, 1994b), presumably because the intimate contact of the ribosomes with the insertion machinery in the membrane prevents their exposure to the chaperone. The only translation product that is a natural substrate of mtHsp70 is Var1, which is the only nonmembrane protein synthesized in the matrix (Herrmann et al, 1994b). The observation that Mba1 and the C-terminus of Oxa1 are critical to position the ribosomes to the insertion sites in the membrane inspired us to test whether the absence of these ribosome-binding proteins makes newly synthesized translation products accessible to mtHsp70. Translation products were radiolabeled in mitochondria containing or lacking Mba1 and/or the ribosome-binding domain of Oxa1. The mitochondria were lysed in the presence of apyrase to deplete the extract of ATP and thereby stabilize the binding of mtHsp70 to its substrates. Then, the radiolabeled translation products that are in contact with mtHsp70 were immunoprecipitated with Hsp70-specific antibodies (Figure 5). Upon deletion of Mba1 or the C-terminus of Oxa1, considerable amounts of Cox1 were found in association with mtHsp70. Cox1 is a protein with 12 transmembrane spans and exhibits the highest overall hydrophobicity of all mitochondrially encoded proteins. Upon deletion of both Mba1 and the C-terminus of Oxa1, a considerable proportion of all translation products visible on our gel system were bound to mtHsp70. Noteworthy, the precursor form of Cox2 that accumulated in the double mutant was efficiently recovered with mtHsp70 (Figure 5, arrowhead). We conclude that Mba1 and the C-terminal ribosome-binding domain of Oxa1 prevent the exposure of nascent chains to soluble proteins of the mitochondrial matrix like mtHsp70. This again points to a critical function of Mba1 and Oxa1 in the cotranslational insertion of membrane proteins in mitochondria.
Figure 5.
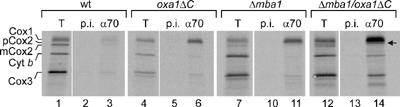
In the absence of Mba1 and the ribosome-binding domain of Oxa1 mitochondrial translation products interact with mtHsp70. Translation products were radiolabeled for 20 min at 37°C in mitochondria from the strains indicated. The mitochondria were reisolated, treated with apyrase to deplete ATP and lysed in 0.1% Triton X-100, 150 mM NaCl, 4 mM EDTA, 20 mM Tris/HCl, pH 7.4, for 10 min at 4°C. The extract was cleared by centrifugation at 17 500 g for 10 min and used for immunoprecipitation with antibodies against mtHsp70 (α70) or preimmune serum (p.i.). Bound proteins were eluted with 1% SDS and analyzed by SDS–PAGE and autoradiography.
Mitochondrial ribosomes remain membrane-associated in the absence of both Mba1 and Oxa1
In contrast to cytosolic ribosomes, mitochondrial ribosomes are tightly associated with membranes and, at least in fungi, can only be released by treatment with detergents (Bunn et al, 1970; Borst and Grivell, 1971). The molecular nature of the membrane binding is unclear. We therefore tested by fractionation on flotation gradients whether the simultaneous deletion of both Mba1 and the C-terminus of Oxa1 caused the release of ribosomes from membranes (Figure 6A). However, ribosomes remained tightly associated with the floating membrane fractions even in Δmba1/oxa1ΔC double mutants. Upon addition of puromycin, a fraction of Mrpl36 still was associated to membranes, even in the Δmba1/oxa1ΔC mitochondria, although there the binding was significantly reduced. By addition of detergent, the ribosomes remained entirely in the bottom fractions (Figure 6B). Thus, Mba1, Oxa1 and nascent chains contribute to the binding of the ribosome to the inner membrane. However, the observation that mitochondrial ribosomes remain partially membrane-associated in the absence of nascent chains, Mba1 and the C-terminus of Oxa1 suggests that additional factors serve as membrane anchors. This is further supported by the observation that membrane binding of mitochondrial ribosomes is highly resistant to salt, whereas the binding of the ribosomes to Mba1 and Oxa1 is sensitive to salt concentrations of higher than 150 mM KCl (Szyrach et al, 2003 and not shown).
Figure 6.
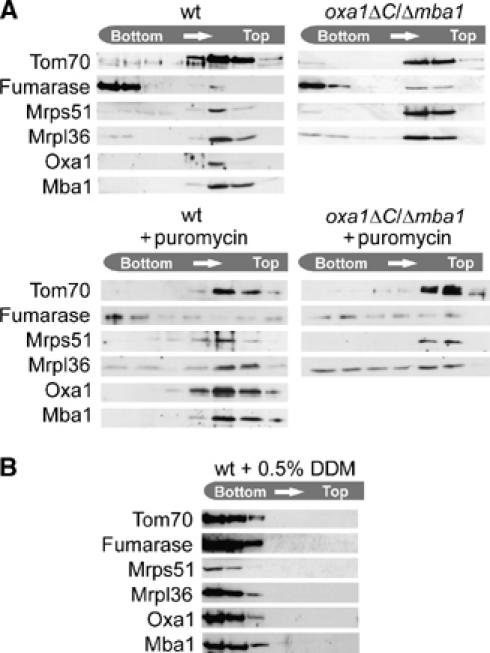
Mitochondrial ribosomes remain associated with the inner membrane in the absence of the ribosome-binding domain of Oxa1 and of Mba1. (A) Mitochondria from the strains indicated were incubated with or without puromycin and disintegrated by freeze thawing in 150 mM KCl, 5 mM EDTA, 20 mM Tris/HCl, pH 7.4. Then, the suspension was adjusted to 2 M sucrose and layers of 2 M sucrose, 1.5 M sucrose, 1 M sucrose in 60 mM KCl, 20 mM Tris/HCl, pH 7.4, were placed on top. After centrifugation at 215 600 g for 16 h at 2°C, the gradient was fractionated. Proteins in the fractions were precipitated by the addition of 12% trichloroacetic acid and analyzed by Western blotting with antibodies against the proteins indicated. (B) Isolated wild-type mitochondria were lysed in 0.5% dodecyl maltoside (DDM), 5 mM EDTA, 20 mM Tris/HCl, pH 7.4, for 15 min at 4°C and processed as in (A).
Suppressors of mba1 and oxa1 mutants
The binding of ribosomes to membranes even in the absence of Mba1 and Oxa1 suggested that additional membrane components might contribute to the membrane binding. Since overexpression of these components might mitigate the defects in oxa1/mba1 double mutants, we performed a multicopy suppressor screen in order to identify such components. We used a mutant lacking the MBA1 gene and harboring a temperature-sensitive oxa1 allele. Both single mutations allowed growth on nonfermentable carbon sources at moderate growth temperatures, whereas the double mutant was unable to respire at all temperatures and showed an impaired growth even on glucose (Preuss et al, 2001). This double mutant was transformed with a highly representative high-copy yeast genomic library, which has successfully been used to clone various genes, including OMS1 and HAP4, two high-copy suppressors of oxa1 partial mutants (Lemaire et al, 2004; Hlavacek et al, 2005). Among 30 000 transformants, 48 colonies showing a plasmid-dependent glycerol growth were obtained and analyzed at the molecular level. In all, 16 of the transforming plasmids carried the MBA1 gene, whereas 32 contained the OXA1 gene, either full length or slightly truncated. At least 10 different overlapping inserts carrying OXA1 were recovered. Additional suppressors were not obtained. Moreover, we found that plasmids carrying either OMS1 or HAP4 could not suppress the growth defect of the mba1 oxa1 double mutant (cf. Supplementary Figure S1). These data suggest that, together, Oxa1 and Mba1 play a highly specific and unique function, which cannot be taken over by other mitochondrial factors. This specific function appears to be the coordination of the cotranslational membrane insertion of mitochondrial translation products presumably by the positioning of the polypeptide exit site of the mitochondrial ribosome to the sites of protein insertion in the inner membrane of mitochondria.
Discussion
The mitochondrial genome encodes exclusively (animals) or predominantly (plants, fungi) hydrophobic membrane proteins. The reduction of the number of hydrophilic translation products during evolution presumably caused the specialization of the mitochondrial protein translation machinery on the production of hydrophobic membrane proteins. As a consequence, protein synthesis in mitochondria appears to occur exclusively or primarily at the matrix face of the inner membrane (Fox, 1996; Grivell et al, 1999; Fiori et al, 2003). We show here that the inner membrane protein Mba1 binds to the large subunit of mitochondrial ribosomes. Our observations suggest that Mba1 serves as a membrane receptor for mitochondrial ribosomes that, in a concerted action with Oxa1, positions the ribosome to the protein insertion site at the inner membrane. Nascent chains are in direct proximity to Mba1 as they can be efficiently crosslinked to Mba1 very early during their synthesis (Preuss et al, 2001). The deletion of Mba1 and of the C-terminus of Oxa1 led to severe defects in the membrane insertion of mitochondrial translation products, whereas single mutants were only moderately affected. This synthetic defect indicates an overlapping and cooperative function of Mba1 and Oxa1 in the membrane recruitment of mitochondrial ribosomes in which the binding of one of both components is still sufficient to coordinate the insertion of mitochondrial translation products. However, deletion of both membrane-bound interaction partners largely impedes productive membrane insertion of translation products, thereby causing a respiration-deficient phenotype. Comparable levels of Mba1 and ribosomes are supported by their highly similar patterns of expression: the transcription levels of Mba1 and ribosomal proteins are surprisingly similar under a large variety of growth conditions (Hughes et al, 2000). Among the nine genes that show the highest similarity in their expression patterns to Mba1, eight encode ribosomal proteins (Table I).
Table 1.
List of proteins the expression of which is closely coregulated with that of Mba1 (Hughes et al, 2000)
| Protein | Function | Prokaryotic family | Corr. coeff.a |
|---|---|---|---|
| Mba1 | 1.0 | ||
| Mrps9 | Protein of mitochondrial ribosome | S9 | 0.741 |
| Mrpl24 | Protein of mitochondrial ribosome | L28 | 0.740 |
| Mam33 | Mitochondrial protein, function unclear | 0.736 | |
| Mrpl6 | Protein of mitochondrial ribosome | L6 | 0.721 |
| YDR115w | Protein of mitochondrial ribosome | L34 | 0.718 |
| Mrp21 | Protein of mitochondrial ribosome | S21 | 0.715 |
| Mnp1 | Protein of mitochondrial ribosome | L7/L12 | 0.713 |
| Mrps28 | Protein of mitochondrial ribosome | S15 | 0.711 |
| Mrpl9 |
Protein of mitochondrial ribosome |
L3 |
0.703 |
| aCorrelation coefficient: see Hughes et al (2000) for details. | |||
| The list is sorted by decreasing correlation coefficients. | |||
Mba1 might be permanently associated with ribosomes as this binding does neither require the presence of nascent chains nor of Oxa1. Mba1 has homologs throughout fungi. Interestingly, database searches for related proteins in animals reveal the sequences of the mitochondrial ribosomal L45 proteins (Mrpl45) as those of highest similarity to the Mba1 family (not shown). Nothing is known on the function of these proteins besides that they were copurified with the large subunit of the mitochondrial ribosome and that they are ubiquitously present in animals but lacking in fungi (Koc et al, 2001). It is conceivable that this Mprl45 group is the functional homolog of the Mba1 family in animals, but experimental evidence for this is missing.
Mba1 is not the only membrane protein that interacts with mitochondrial ribosomes. At least in fungi, the tight membrane association of mitochondrial ribosomes appears to rely on multiple interactions with components of the inner membrane as depicted in Figure 7: (1) mitochondrial mRNAs were shown to be associated to the inner membrane via membrane-associated translational activators (Costanzo and Fox, 1988; Fox, 1996). (2) Oxa1 (Jia et al, 2003; Szyrach et al, 2003) and Mba1 (this study) bind to the large subunit of the ribosome and thereby tether it to the inner membrane. (3) Both subunits of the mitochondrial ribosome apparently remain at least partially membrane bound even in the absence of Mba1 and Oxa1, indicating that ribosomes are connected to the membrane via additional linkages (this study). Inner membrane proteins like Mdm38 and Cox11 might represent such tethering factors as both membrane proteins were recently shown to bind to mitochondrial ribosomes (Khalimonchuk et al, 2005; Frazier et al, 2006). The tight association of fungal mitochondrial ribosomes with the inner membrane is known for a long time as the addition of detergent is necessary for their isolation (Bunn et al, 1970; Borst and Grivell, 1971). A similar association to the inner membrane was reported for animal mitochondria (Liu and Spremulli, 2000). In a recent study, the ribosomal protein Mrpl32 (a homolog of the bacterial L32 protein) was reported to become tightly associated with the inner membrane following its processing by Yta10 and Yta12, the subunits of the m-AAA protease of the inner membrane (Nolden et al, 2005). While the molecular basis for this membrane association is still unclear, this observation indicates a critical function of mitochondrial proteins in membrane binding, which is under the control of the m-AAA protease. Interestingly, overexpression of Oxa1 or Mba1 suppresses the defects of yta10 and yta12 mutants (Rep et al, 1996) potentially because high levels of Oxa1 or Mba1 lead to an increased membrane recruitment of ribosomes, which bypasses the need of the Mrpl32-dependent membrane binding.
Figure 7.
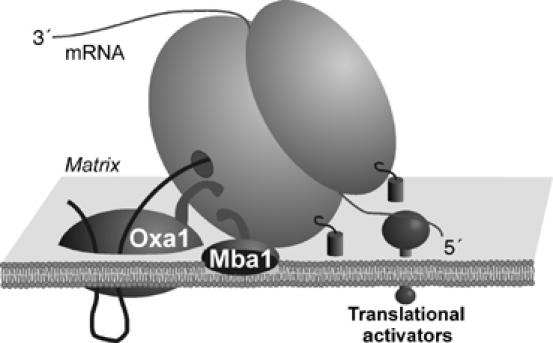
Model for membrane interaction of mitochondrial ribosomes. Mitochondrial ribosomes are tightly associated with the inner membrane. This interaction appears to rely on at least three mechanisms: (1) mitochondrial mRNAs are bound to the inner membrane by translational activators. (2) The membrane proteins Mba1 and Oxa1 are associated with the large subunit of the mitochondrial ribosome. The results shown in this study indicate that Mba1 and the C-terminal ribosome-binding domain of Oxa1 are critical to achieve close contact of the polypeptide exit tunnel of the ribosome and the insertion machinery of the inner membrane. (3) Additional factors, like Mdm38 or Cox11, contribute to the membrane binding of ribosomes independently of nascent chains, Mba1 or Oxa1.
The observation that mitochondrial ribosomes are tethered to the membrane even in the absence of Oxa1 and Mba1 raises the question why translational activators and the binding by Oxa1 and Mba1 are at all necessary for proper membrane insertion of translation products. It was proposed that translational activators might recruit ribosomes to specific locations in the inner membrane, which are dedicated for the insertion and assembly of membrane proteins (Naithani et al, 2003). Such a topological segregation of the inner membrane into regions of different physiological functions is supported by both biochemical and electron microscopic evidence (for a review see Frey and Mannella, 2000). In addition, Mba1 and the ribosome-binding domain of Oxa1 are apparently required to bring the polypeptide exit site of the ribosome in close proximity to the insertion machinery of the inner membrane (Jia et al, 2003; Szyrach et al, 2003 and this study). Owing to the intimate contact of ribosomes to the membrane, mitochondrial chaperones like mtHsp70 do not have access to nascent membrane proteins (Herrmann et al, 1994b). Interestingly, in the absence of Mba1 and the ribosome-binding domain of Oxa1, nascent chains are not properly delivered from the ribosome to the membrane and become exposed to mtHsp70. The binding of chaperones might prevent that the translation products form aggregates, which were not found in Δmba1/oxa1ΔC mutants (not shown). Nevertheless, in the absence of Mba1 and the ribosome-binding domain of Oxa1, translation products fail to be inserted into the membrane pointing at an obligate coupling of the synthesis and membrane insertion of mitochondrially encoded proteins. It is conceivable that the progression of a nascent chain through the ribosome tunnel is used to energetically drive its translocation across the membrane. However, such a ‘ribosome pushing effect' was even not shown unequivocally for the much better studied translocation of nascent chains across the membrane of the endoplasmic reticulum. It will be an exciting task in the future to analyze the energetic basis of the translocation of newly synthesized membrane proteins in mitochondria.
Materials and methods
Yeast strains and growth media
All strains used in this study were isogenic to the wild-type strain W303-1A. For generation of the mba1 deletion mutants, the entire coding region of the MBA1 gene was replaced by a HIS3 cassette. The generation of the oxa1ΔC mutant (Oxa11−331) lacking residues 332–402 (Szyrach et al, 2003) and the Δmba1/oxa1ts double mutant used for the genetic screen was described before (Preuss et al, 2001). For all experiments in this study with truncated Oxa1 mutants, the Oxa11−331 variant was used. Yeast cultures were grown at 30°C in lactate medium, YP medium supplemented with 2% galactose or minimal medium supplemented with 20 μg/ml adenine, histidine and tryptophan, and 30 μg/ml of leucine and lysine (Sherman et al, 1986; Herrmann et al, 1994a). Mitochondria were isolated as described previously (Herrmann et al, 1994a).
Genetic screen for multicopy suppressors of mba1/oxa1 mutants
The mba1/oxa1ts double mutant strain (HHY0064) was transformed with a high-copy yeast genomic library cloned in the 2 μm URA3 vector pFL44L (Stettler et al, 1993). Transformants were selected on minimal medium lacking uracil, and replica plated onto two plates of glycerol medium, one incubated at 28°C and the other at 36°C. Plates were followed for 2 weeks, and positive colonies (at 28°C and/or 36°C) were patched on glycerol medium and tested again for growth at 28 and 36°C. Plasmids containing OXA1 were expected to complement the mutant at both temperatures, whereas expression of MBA1 should permit growth only at 28°C. To identify the inserts, plasmids were analyzed on the basis of restriction patterns and PCR with MBA1-, OXA1- and vector-specific primers. A control transformation with the empty vector did not yield any [Gly+] colony within 7000 transformants. In addition, the library allowed easy cloning of the ADE2 gene (one positive colony in 2300 transformants).
In vitro binding experiments
The entire open-reading frame of MBA1 was cloned into the BamHI and HindIII sites of pMalcRI (New England Biolabs) to allow expression of an MBP-Mba1 fusion protein in the E. coli BL21(DE3) strain (Stratagene). The fusion protein, or recombinant MBP for control, was purified and immobilized on amylose resin. Mitochondrial translation products were radiolabeled in isolated mitochondria. The mitochondria were washed and lysed in 1% Triton X-100, 50 mM KCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 20 mM HEPES, pH 7.4. The resulting lysate was cleared by centrifugation for 15 min at 50 000 g at 4°C and incubated with the immobilized recombinant protein for 1 h at 25°C. The resin was washed extensively. Bound proteins were eluted with SDS-containing sample buffer and analyzed by immunoblotting.
Fractionation of mitochondrial ribosomes on sucrose gradients
Isolated mitochondria (1 mg) were lysed in 1% Triton X-100, 25 mM KCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 20 mM HEPES, pH 7.4, for 30 min at 4°C. After a clarifying spin for 15 min at 30 000 g at 2°C, the extract was layered onto a continuous 4 ml sucrose gradient (0.3–1 M sucrose in 1% Triton X-100, 25 mM KCl, 5 mM EDTA, 20 mM HEPES, pH 7.4) and centrifuged for 75 min at 485 000 g in a Beckman SW60 Ti rotor at 4°C. Fractions were collected and containing proteins were precipitated with 12% trichloroacetic acid. The resulting pellets were dissolved in sample buffer, separated by SDS–PAGE and analyzed by immunoblotting.
Subfractionation of mitochondrial translation products
Following radiolabeling of mitochondrial translation products, mitochondria were washed and incubated with extraction buffer (0.1 M sodium carbonate, 4.5 M urea) for 30 min at 25°C. The lysate was adjusted to 1 ml 1.6 M sucrose in extraction buffer, transferred into a centrifugation tube and layers of 2 ml 1.4 M sucrose and 1 ml 0.25 M sucrose in extraction buffer were put on top. The tubes were centrifuged in a SW60 Ti rotor for 4 h at 337 000 g. Fractions were collected and proteins were precipitated with 12% trichloroacetic acid. The resulting pellets were dissolved in sample buffer and analyzed by immunoblotting and autoradiography.
Miscellaneous
The radiolabeling of translation products in mitochondria (Herrmann et al, 1994a) and the analysis of their interactions with mtHsp70 by immunoprecipitation was assessed as described (Herrmann et al, 1994b).
Supplementary Material
Supplementary Figure S1
Acknowledgments
We are grateful to Sandra Esser for excellent technical assistance and to Gregor Szyrach, Michael Horn and Martin Schmidt for help with some experiments. We thank Walter Neupert for stimulating discussions and comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB Teilprojekt B05 to JMH) and by an EMBO long-term fellowship to SF.
References
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T (1996) The YTA10–12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85: 875–885 [DOI] [PubMed] [Google Scholar]
- Bauer M, Behrens M, Esser K, Michaelis G, Pratje E (1994) PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol Gen Genet 245: 272–278 [DOI] [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science 278: 2123–2126 [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Chalvet F, Hamel P, Slominski PP, Dujardin G (1994) OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol 239: 201–212 [DOI] [PubMed] [Google Scholar]
- Borst P, Grivell LA (1971) Mitochondrial ribosomes. FEBS Lett 13: 73–88 [DOI] [PubMed] [Google Scholar]
- Borst P, Grivell LA (1978) The mitochondrial genome of yeast. Cell 15: 705–723 [DOI] [PubMed] [Google Scholar]
- Bunn CL, Mitchell CH, Lukins HB, Linnane AW (1970) Biogenesis of mitochondria. 18. A new class of cytoplasmically determined antibiotic resistant mutants in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 67: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Perea J, Shu YM, Samatey FA, Popot JL, Jacq C (1995) Limitations to in vivo import of hydrophobic proteins into yeast mitochondria—the case of a cytoplasmically synthesized apocytochrome b. Eur J Biochem 228: 762–771 [PubMed] [Google Scholar]
- Costanzo MC, Fox TD (1988) Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc Natl Acad Sci USA 85: 2677–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori A, Mason TL, Fox TD (2003) Evidence that synthesis of the Saccharomyces cerevisiae mitochondrially encoded ribosomal protein Var1p may be membrane localized. Eukaryot Cell 2: 651–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TD (1996) Genetics of mitochondrial translation. In Translational Control Hershey JWB, Matthews MB, Sonnenberg N (eds) pp 733–758. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P (2006) Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol 172: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey TG, Mannella CA (2000) The internal structure of mitochondria. Trends Biochem Sci 25: 319–324 [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G (1983) Transient involvement of signal recognition particle and its receptor in the microsomal membrane prior to protein translocation. Cell 35: 677–685 [DOI] [PubMed] [Google Scholar]
- Glick BS, von Heijne G (1996) Saccharomyces cerevisiae mitochondria lack a bacterial-type Sec machinery. Protein Sci 5: 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green-Willms NS, Fox TD, Costanzo MC (1998) Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol Cell Biol 18: 1826–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell LA, Artal-Sanz M, Hakkaart G, de Jong L, Nijtmans LG, van Oosterum K, Siep M, van der Spek H (1999) Mitochondrial assembly in yeast. FEBS Lett 452: 57–60 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hell K, Herrmann J, Pratje E, Neupert W, Stuart RA (1997) Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett 418: 367–370 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Fölsch H, Neupert W, Stuart RA (1994a) Isolation of yeast mitochondria and study of mitochondrial protein translation. In Cell Biology: A Laboratory Handbook Celis JE (ed), Vol. 1, pp. 538–544. San Diego: Academic Press [Google Scholar]
- Herrmann JM, Neupert W (2003) Protein insertion into the inner membrane of mitochondria. IUBMB Life 55: 219–225 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W (1994b) Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol 127: 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavacek O, Bourens M, Salone V, Lachacinski N, Lemaire C, Dujardin G (2005) The transcriptional activator HAP4 is a high copy suppressor of an oxa1 yeast mutation. Gene 354: 53–57 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH (2000) Functional discovery via a compendium of expression profiles. Cell 102: 109–126 [DOI] [PubMed] [Google Scholar]
- Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA (2003) Yeast Oxa1 interacts with mitochondrial ribosomes: The importance of the C-terminal hydrophilic region of Oxa1. EMBO J 22: 6438–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Ostermann K, Rödel G (2005) Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr Genet 47: 223–233 [DOI] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL (2001) The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276: 43958–43969 [DOI] [PubMed] [Google Scholar]
- Kuhn A, Stuart R, Henry R, Dalbey RE (2003) The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol 13: 510–516 [DOI] [PubMed] [Google Scholar]
- Lemaire C, Guibet-Grandmougin F, Angles D, Dujardin G, Bonnefoy N (2004) A yeast mitochondrial membrane methyltransferase-like protein can compensate for oxa1 mutations. J Biol Chem 279: 47464–47472 [DOI] [PubMed] [Google Scholar]
- Liu M, Spremulli L (2000) Interaction of mammalian mitochondrial ribosomes with the inner membrane. J Biol Chem 275: 29400–29406 [DOI] [PubMed] [Google Scholar]
- Ménétret JF, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW (2000) The structure of ribosome–channel complexes engaged in protein translocation. Mol Cell 6: 1219–1232 [DOI] [PubMed] [Google Scholar]
- Naithani S, Saracco SA, Butler CA, Fox TD (2003) Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell 14: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123: 277–289 [DOI] [PubMed] [Google Scholar]
- Pajic A, Tauer R, Feldmann H, Neupert W, Langer T (1994) Yta10p is required for the ATP-dependent degradation of polypeptides in the inner membrane of mitochondria. FEBS Lett 353: 201–206 [DOI] [PubMed] [Google Scholar]
- Preuss M, Leonhard K, Hell K, Stuart RA, Neupert W, Herrmann JM (2001) Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J Cell Biol 153: 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M, Ott M, Funes S, Luirink J, Herrmann JM (2005) Evolution of mitochondrial Oxa proteins from bacterial YidC: inherited and acquired functions of a conserved insertion machinery. J Biol Chem 280: 13004–13011 [DOI] [PubMed] [Google Scholar]
- Rep M, Grivell LA (1996) MBA1 encodes a mitochondrial membrane-associated protein required for biogenesis of the respiratory chain. FEBS Lett 388: 185–188 [DOI] [PubMed] [Google Scholar]
- Rep M, Nooy J, Guélin E, Grivell LA (1996) Three genes for mitochondrial proteins suppress null-mutations in both AFG3 and RCA1 when overexpressed. Curr Genet 30: 206–211 [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks J (1986) Methods in Yeast Genetics: A Laboratory Course. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P (1993) A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet 239: 169–176 [DOI] [PubMed] [Google Scholar]
- Stuart RA (2002) Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochem Biophys Acta 1592: 79–87 [DOI] [PubMed] [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM (2003) Ribosome binding to the Oxa1 complex facilitates cotranslational protein insertion in mitochondria. EMBO J 22: 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Myers AM (1986) Genetics of mitochondrial biogenesis. Annu Rev Biochem 55: 249–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1


