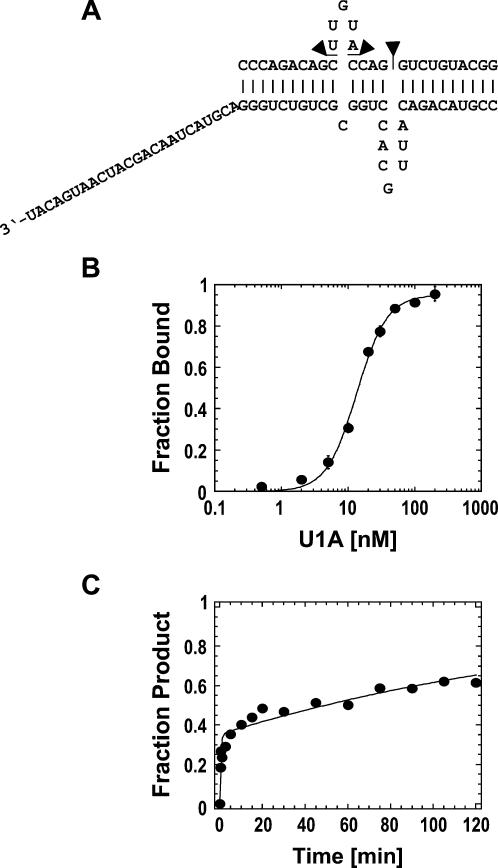
FIGURE 7.
Altered U1A-based RNP. (A) RNP design. Three nucleotides were deleted from the upper RNA strand (indicated by triangles) of the RNA complex with the authentic U1A binding site (Fig. 4A). (B) Equilibrium binding of U1A to the altered RNA complex. Data points represent the average of three independent measurements. Data were fit to the Hill-equation (K D = 13.5 ± 0.7 nM, n = 1.9 ± 0.2). (C) Spontaneous dissociation of U1A from the RNA. The representative time course was fit to the sum of two exponentials. Dissociation rate constants were, for the first phase: k I d = 2.2 ± 0.6 min−1, and for the second phase: k II d = (5.3 ± 0.7) × 10−3 min−1.