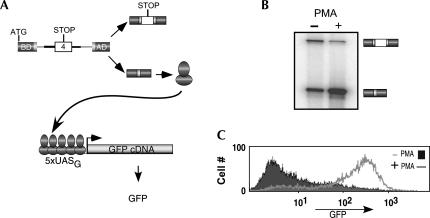
FIGURE 1.
Design of a two-component splicing reporter that makes use of transcriptional synergy to augment detection of alternative splicing. (A) Schematic of the SC-VP16 (top) and pG5-GFP (bottom) constructs that were transfected into the JSL1 cell line. For SC-VP16 the dark gray boxes represent the coding sequence for the Gal4 DNA binding domain (BD) and VP16 transcription activation domain (AD), the light gray sequences derive from the human β-globin gene and the white box is variable exon 4 from the CD45 gene. For pG5-GFP the five binding sites of Gal 4 (5xUASg) are shown binding to the Gal4-VP16 fusion protein and drive expression of the GFP cDNA. (B) RT-PCR assay done under low-cycle conditions determined empirically to yield a quantitative assessment of the change in the ratio of splicing products from the pSC-VP16 between resting (−PMA) and stimulated (+PMA) conditions. Exon 4 inclusion (top band) to exclusion (bottom band) ratio in resting cells is 0.9 ± 0.1, though this ratio in activated cells is 0.2 ± 0.05. (C) Flow cytometry analysis of GFP expression in resting (filled) or stimulated (line) cells. Cells growing at a similar density were washed in PBS and analyzed after gating on live cells.