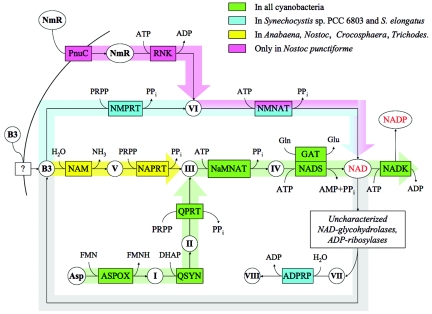
FIG. 1.
Biochemical transformations (arrows) directly involved in the biosynthesis, salvage, and recycling of NAD(P) in cyanobacteria. Functional roles in this subsystem (mostly enzymes) are shown by abbreviations in boxes. ADPRP, ADP-ribose pyrophosphatase (EC 3.6.1.13); ASPOX, l-aspartate oxidase (EC 1.4.3.16); GAT, glutamine amidotransferase chain of NAD synthetase; NADK, NAD kinase (EC 2.7.1.23); NADS, NAD synthetase (EC 6.3.1.5); NAM, nicotinamidase (EC 3.5.1.19); NaMNAT, nicotinate-nucleotide adenylyltransferase (EC 2.7.7.18); NAPRT, nicotinate phosphoribosyltransferase (EC 2.4.2.11); NMNAT, nicotinamide-nucleotide adenylyltransferase (EC 2.7.7.1); NMPRT, nicotinamide phosphoribosyltransferase (EC 2.4.2.12); PnuC, ribosyl nicotinamide transporter, PnuC-like; QPRT, quinolinate phosphoribosyltransferase (decarboxylating) (EC 2.4.2.19); QSYN, quinolinate synthetase (EC 4.1.99.-); RNK, ribosylnicotinamide kinase (EC 2.7.1.22). Key metabolites (precursors and products) and intermediates are shown by abbreviations or roman numerals in circles. Asp, l-aspartate; B3, nicotinamide, vitamin B3; NADP, NAD phosphate; NmR, N-ribosylnicotinamide; I, iminoaspartate; II, quinolinic acid; III, nicotinate mononucleotide (NaMN); IV, deamido-NAD; V, nicotinic acid; VI, nicotinamide mononucleotide (NMN); VII, ADP-ribose; VIII, ribose phosphate. Other metabolites are shown by standard symbols and abbreviations: DHAP, dihydroxyacetone phosphate; FMN, flavin mononucleotide; FMNH, flavin mononucleotide, reduced; PRPP, 5-phosphoribosyl-1-pyrophosphate; PPi, pyrophosphate. Thick bars with arrows outline individual pathways; the coloring scheme reflects the occurrence of respective genes and corresponding pathways in various cyanobacteria, as shown in the inset. For example, the green color indicates genes and pathways (de novo and universal) present in all compared cyanobacteria.