FIG. 6.
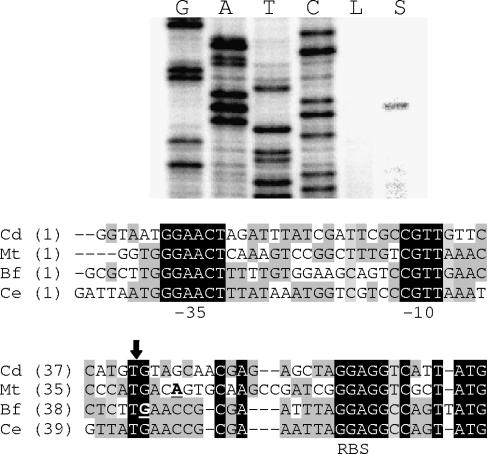
The gel image includes the results of primer extension assays. The primer sigBPE3 was used to generate both the sequencing ladder (G, A, T, C) and the primer extension products. The lanes containing primer extension reaction mixtures are labeled L for template RNA isolated from low-iron cultures and S for template RNA isolated from low-iron cultures containing SDS. The DNA sequences of the regions upstream of sigB in C. diphtheriae (Cd), M. tuberculosis (Mt), B. flavum (Bf), and Corynebacterium efficiens (Ce) are shown below the image of the gel. The T residue corresponding to the start of the sigB transcript in C. diphtheriae is indicated by the vertical arrow. The underlined boldface bases in the Mt and Bf sequences indicate the previously reported transcript starts in those bacteria. The sequences corresponding to −10 and −35 sequences used by ECF sigma factors σE and σH are labeled. The proposed ribosome binding site is labeled RBS.