Abstract
Several species of mycoplasmas rely on an unknown mechanism to glide across solid surfaces in the direction of a membrane protrusion at the cell pole. Our recent studies on the fastest species, Mycoplasma mobile, suggested that a 349-kDa protein, Gli349, localized at the base of the membrane protrusion called the neck, forms legs that stick out from the neck and propel the cell by repeatedly binding to and releasing from a solid surface, based on the energy of ATP hydrolysis. Here, the Gli349 protein was isolated from mycoplasma cells and its structure was analyzed. Gel filtration analysis showed that the isolated Gli349 protein is monomeric. Rotary shadowing electron microscopy revealed that the molecular structure resembles the symbol for an eighth note in music. It contains an oval foot 14 nm long in axis. From this foot extend three rods in tandem of 43, 20, and 20 nm, in that order. The hinge connecting the first and second rods is flexible, while the next hinge has a distinct preference in its angle, near 90 degrees. Molecular images revealed that a monoclonal antibody that can bind to the position at one-third of the total peptide length from the N terminus bound to a position two-thirds from the foot end, suggesting that the foot corresponds to the C-terminal region. The amino acid sequence was assigned to the molecular image, and the topology of the molecule in the gliding machinery is discussed.
Mycoplasmas are commensal and occasionally parasitic bacteria that have small genomes and lack a peptidoglycan layer (19). Several Mycoplasma species form membrane protrusions, such as the head-like structure in Mycoplasma mobile and the attachment organelle in Mycoplasma pneumoniae (11, 14, 21, 23, 25). On solid surfaces, these species exhibit gliding motility in the direction of the protrusion; this motility is believed to be involved in the pathogenicity of mycoplasmas (3, 10, 14). Interestingly, mycoplasmas have no surface flagella or pili, and their genomes contain no known genes related to bacterial motility. In addition, no homologs of motor proteins that are common in eukaryotic motility have been found (8, 14).
M. mobile, isolated from the gills of a freshwater fish in the early 1980s, is the fastest-gliding mycoplasma. It glides smoothly and continuously on glass at an average speed of 2.0 to 4.5 μm/s, or three to seven times the length of the cell per s, exerting a force of up to 27 pN (6, 16, 17, 20). Recently, three huge proteins—Gli123 (123 kDa), Gli349 (349 kDa), and Gli521 (521 kDa)—were found to be clustered at the neck; they are responsible for the positioning of other gliding proteins, binding to glass during gliding, and generating and/or transmitting force, respectively (12, 24, 26, 28).
Rapid-freeze and freeze fracture rotary shadowing electron microscopy (EM) revealed that many spike-like structures 50 nm in length stick out around the neck and bind to the glass surface with their distal ends (15). The spike seems to be composed of a Gli349 molecule and to function as a leg in the gliding mechanism. Analyses of a Triton model, designated the “ghost,” where the cell membrane is permeabilized and intracellular reactions can be manipulated externally, demonstrated that the gliding mechanism is driven by the energy of ATP (7, 27). These observations led us to assume that cells are propelled by spikes composed of Gli349 that repeatedly bind to and release from the glass, driven by the force exerted from or through the Gli521 molecule, based on the energy of ATP hydrolysis (4, 14, 24, 26). This putative mechanism may be applied to M. pneumoniae, although protein P1, responsible for glass binding, does not share a primary structure with Gli349 of M. mobile, because a monoclonal antibody against P1 affects the gliding of M. pneumoniae in a very similar way to an antibody against Gli349 on M. mobile gliding (9, 22).
To obtain more concrete images of the gliding mechanism, it is critical to know the structure of the Gli349 protein of M. mobile. Detailed analyses of the amino acid sequence revealed that about 60% of the length of the Gli349 molecule is composed of 18 repeats, each consisting of about 100 amino acid residues (13). However, neither all nor part of the molecular shape can be predicted, because the amino acid sequence of Gli349 does not show significant similarity to those of proteins other than the ortholog found in Mycoplasma pulmonis, a gliding species closely related to M. mobile. Therefore, experimental analyses are essential in order to understand the structure of Gli349. In the present study, we isolated the Gli349 protein, observed its molecular shape by rotary shadowing EM, and assigned its amino acid sequence to the image of the molecule.
MATERIALS AND METHODS
Strains and culture conditions.
M. mobile strain 165K (ATCC 43663) was grown at 25°C in Aluotto medium, consisting of 2.1% heart infusion broth, 0.56% yeast extract, 10% horse serum, 0.0025% thallium acetate, and 0.005% ampicillin, to an optical density at 600 nm of around 0.1 (1, 18).
Purification of Gli349.
All procedures were done at 4°C. Cells from 1 liter of culture were centrifuged at 14,000 × g for 10 min and washed twice with phosphate-buffered saline consisting of 75 mM sodium phosphate (pH 7.3) and 68 mM NaCl. The cells were suspended to an optical density at 600 nm of 20 in 10 mM Tris-HCl (pH 8.0), 0.1 mM phenylmethylsulfonyl fluoride and then were mixed with Triton X-100 to 1% (vol/vol). After gentle shaking for 1 h, the suspension was ultracentrifuged at 450,000 × g for 30 min (step 1). The supernatant was fractionated by stepwise salting out with ammonium sulfate of 30% and 35% saturation in the same buffer as in step 1.
The insoluble fractions at 35% saturation were recovered by centrifugation at 22,000 × g for 15 min (step 2). The recovered fraction was dissolved and dialyzed overnight by 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 5.9). The insoluble fraction caused by this pH shift was removed by centrifugation at 22,000 × g for 15 min (step 3). The soluble fraction was loaded onto a charged Hi-Trap 1-ml Q Sepharose (Fast Flow) column (GE Healthcare, Milwaukee, WI) with a flow rate of 1.0 ml/min, and equilibrated with 10 mM MES (pH 5.9). The proteins were eluted with a linear gradient from 0 to 1 M NaCl in 10 mM MES (pH 5.9) of a total volume of 36 ml and fractionated into 1.5-ml aliquots. Gli349 was eluted around 0.15 M NaCl (step 4).
The homogeneity of protein fractions was estimated by the densitometry of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels with a scanner (GT-9800F; Epson, Nagano, Japan) and analyzing software, Image-J version 1.33u (National Institutes of Health). The fractions were dialyzed against 10 mM ammonium acetate (pH 6.5) overnight and concentrated using Biomax-10 (Millipore, Bedford, MA) to 0.3 mg/ml. This protein fraction was held at −20°C in 33% (vol/vol) glycerol and 0.3 M ammonium acetate (pH 6.5), if necessary, and used within a month.
Gel filtration assay.
The final Gli349 fraction, containing 0.15 mg protein in 0.5 ml, was applied to a Hi Load 16/60 Superdex 200 pg set (GE Healthcare) on ÄKTA prime (GE Healthcare) and eluted with a buffer consisting of 0.2 M NaCl and 10 mM Tris-HCl, pH 8.0, with a flow rate of 1 ml/min at room temperature. The sample elution was monitored by absorbance at 280 nm. Ferritin, aldolase, and chymotrypsinogen were used as standards.
Rotary shadowing EM.
Gli349 was diluted to 20 μg/ml in 33% (vol/vol) glycerol and 0.3 M ammonium acetate and sprayed on a freshly cleaved mica surface as described previously (2). For the analysis of Gli349 bound by a monoclonal antibody, MAb7 (12), the antibody was purified from the hybridoma supernatant by Hi-Trap Protein G HP (GE Healthcare). Gli349 and MAb7 were mixed to be 20 and 10 μg/ml, respectively, in similar molar amounts, incubated for 1 h at 4°C, and sprayed on a mica surface as mentioned above. The mica was then dried under vacuum, rotary shadowed with platinum at an angle of 8 degrees, and supported with carbon (HUS5-GB; Hitachi, Tokyo, Japan).
The replicas were observed by an H-7000 transmission electron microscope (Hitachi, Tokyo, Japan) at 90 kV. Whole micrographs were digitized as 16-bit images using DuoScan HiD (Agfa, Mortsel, Belgium). Each particle image was picked out by EMAN, version 1.6 (http://ncmi.bcm.tmc.edu/∼stevel/EMAN/doc/). The lengths and angles of molecular images were analyzed by Scion Image PC beta version 4.0.2 (Scion Corp., Frederick, MD) and Adobe Photoshop version 7.0.1 (Adobe, San Jose, CA).
RESULTS
Isolation of Gli349.
Our previous observations, listed below, suggest that the Gli349 molecule is mostly outside of and anchored to the membrane (12, 13, 26). (i) The Gli349 molecule is predicted to have a transmembrane segment at its N-terminal region, based on its amino acid sequence. (ii) A monoclonal antibody can label the Gli349 molecule and inhibit its function from outside the cell. (iii) The treatment of mycoplasma cells with 1% Triton X-100 solubilized 60% of Gli349 from the cells, leaving many proteins insoluble. This assumption leads us to the idea that treatment of cells with a detergent may release the molecules specifically from the cells.
We isolated the Gli349 protein using the four-step procedure presented in Fig. 1. In step 1 the collected cells suspended in buffer were treated with 1% Triton X-100 to solubilize Gli349, followed by the removal of insoluble fractions by ultracentrifugation. In step 2 the fraction including Gli349 was applied to stepwise ammonium sulfate fractionation at 30% and 35% saturation in the presence of Triton X-100. The fraction at 35% saturation was dissolved and applied to the following steps. Gli349 was found also in the fractions at 40% saturation. However, this fraction was not used, because it contained a significant amount of another protein, MvspI, which is not easy to remove by the following procedures.
FIG. 1.
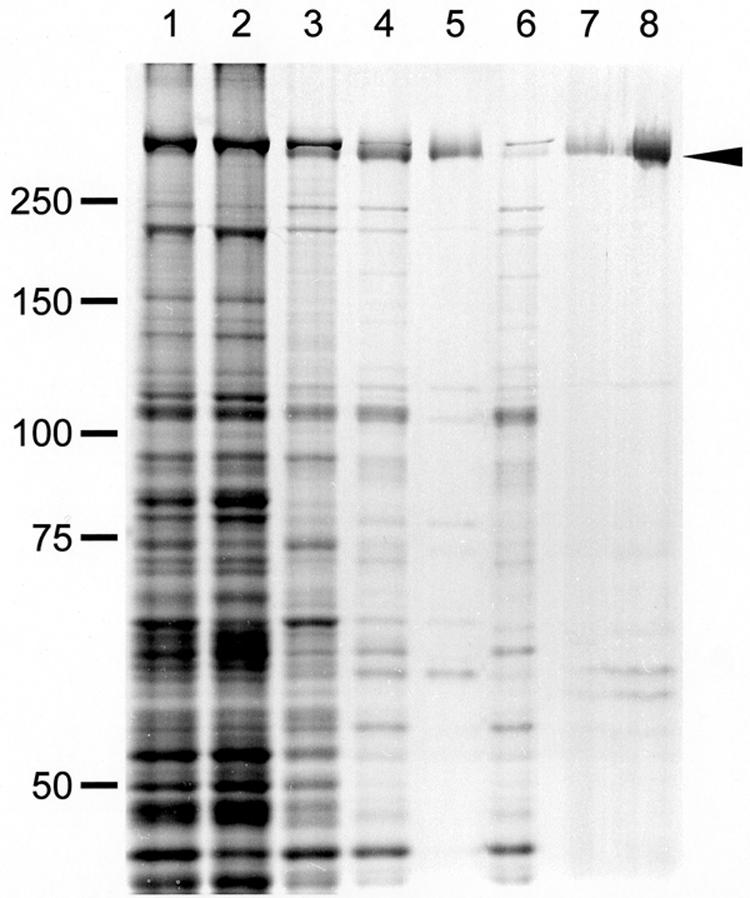
Protein profiles of fractions in the Gli349 purification procedure. Gli349 protein was purified through four steps: (i) Triton X-100 treatment of cells and centrifugation, (ii) stepwise ammonium sulfate fractionation, (iii) precipitation of other proteins by a pH shift from 8.0 to 5.9, and (iv) Q-Sepharose (anion exchanger) column chromatography. The arrowhead shows the Gli349 protein band. Lane 1: whole-cell lysate. Lane 2: Triton-insoluble fraction of step 1. Lane 3: Triton-soluble fraction of step 1. Lane 4: precipitate of 35% saturation ammonium sulfate in step 2. Lane 5: supernatant of step 3. Lane 6: precipitate of step 3. Lane 7: fraction eluted at 0.15 M NaCl in step 4. Lane 8: 10-fold concentration of the fraction in lane 7. Each fraction was subjected to SDS-7.5% PAGE with a 3-mm lane width and stained by the reverse-staining method. Protein fractions derived from 0.2-, 0.5-, and 5-ml cultures were applied to lanes 1 to 6, 7, and 8, respectively. Molecular masses are indicated on the left in kilodaltons.
In step 3 the ammonium sulfate precipitate was dissolved and dialyzed by a buffer of pH 5.9. With this pH shift, 35% of the protein formed an insoluble fraction. This insoluble fraction, which did not include Gli349, was removed by centrifugation. In step 4 the soluble fraction including Gli349 was applied to a column of Q Sepharose, a weak anion exchanger, and eluted with a 0 to 1 M NaCl gradient. Gli349 was found at around 0.15 M NaCl, and the eluate was used for the following experiments. This fraction did not show any other major bands in the gel image of SDS-PAGE even when stained by the reverse-staining method, in which the sensitivity was 10 times that of Coomassie brilliant blue (5). A few contaminating protein bands were found when the eluate was concentrated 10-fold. The homogeneity of the final fraction was estimated as 92.7% from densitometry. Finally, 70 μg of Gli349 was obtained from 1 liter of mycoplasma culture. Assuming that a cell contains 450 molecules of Gli349 (26), the purification yield was calculated to be about 40%.
Molecular weight of isolated Gli349.
To examine the possibility of self-assembly, the isolated Gli349 protein was applied to a gel filtration assay (Fig. 2). The elution pattern showed a single peak, and its molecular mass was calculated as 354 kDa based on the retention time. We concluded that the isolated Gli349 is in a monomer state.
FIG. 2.
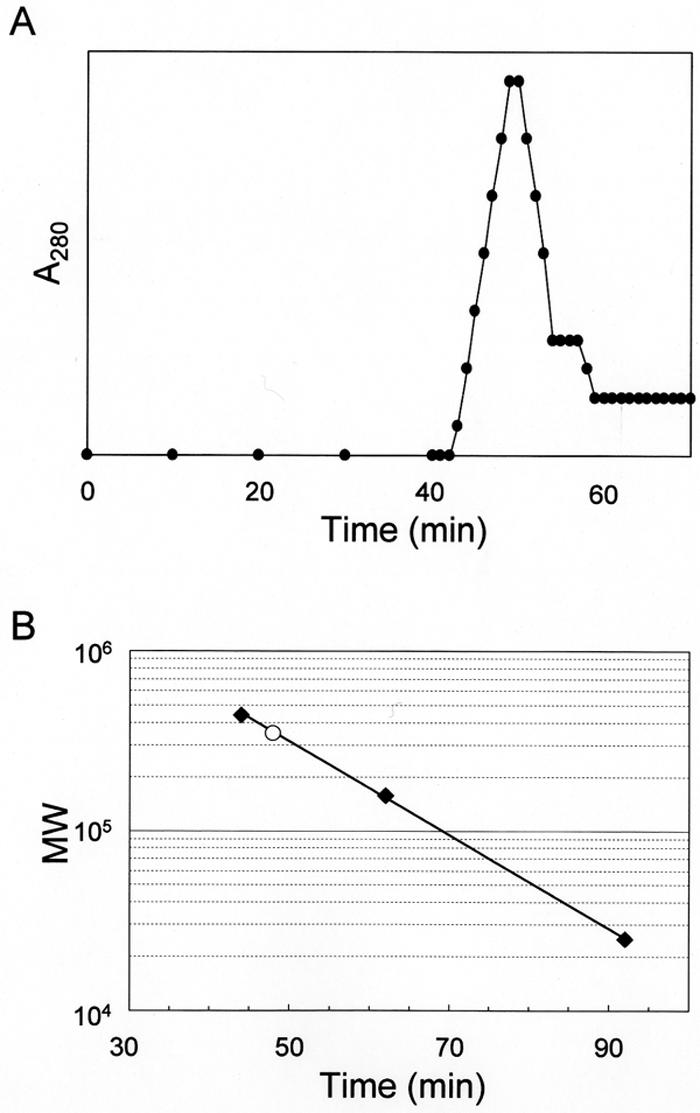
Gel filtration assay of isolated Gli349. (A) Elution pattern of Gli349 protein. (B) Molecular weight of native Gli349 protein. Molecular weight and retention time are presented on the y and x axes, respectively. Solid squares show size standards: ferritin (440 kDa), aldolase (158 kDa), and chymotrypsinogen (25 kDa). The retention time and calculated molecular weight (MW) of Gli349 are presented by the open circle.
Shape and dimensions of Gli349.
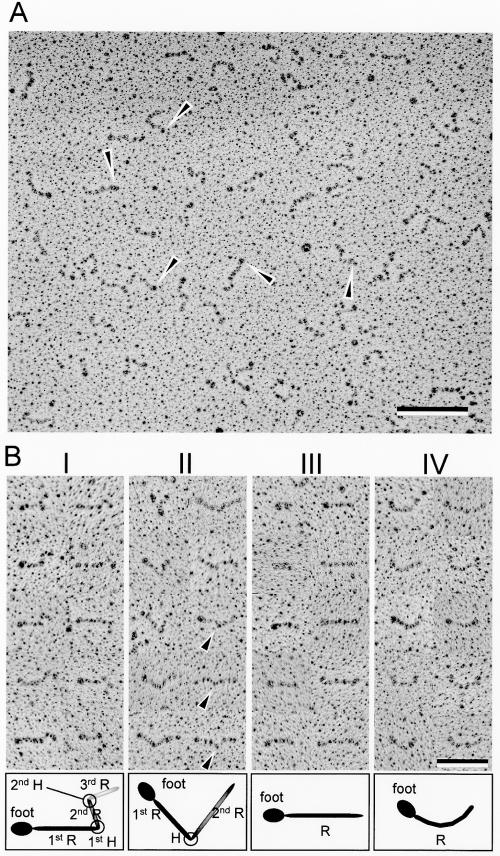
The purified Gli349 protein was rotary shadowed and observed by transmission EM (Fig. 3). Rod-shaped particles were found to have total lengths of from 18.9 to 124.4 nm. These structures were found basically in all fields of a grid, and the frequency of their appearance changed in correspondence with the concentration of Gli349 protein used. These facts suggest that the rod-shaped particles reveal the shape of the Gli349 molecule. Almost all particles had a dense oval part, which we called a foot, at the end of the pole.
FIG. 3.
Rotary-shadowed EM of Gli349 molecules. (A) Image of a field 1,354 nm wide and 1,122 nm high. Arrowheads indicate spherical and thick parts, called feet. Bar, 200 nm. (B) Isolated images and classification into four types. The molecular images were classified into types I, II, III, and IV, featuring two bends, one bend, linear form with no bend, and curved form with no bend, respectively. The images are aligned so that the feet are on the left side. Ten molecular images are presented for each type indicated at the top of each column. A distinct bend was observed in type II images, as marked by triangles, but not in type IV images. The schematics below each image present each part of the image, as the first rod (R), first hinge (H), and so on. Bar, 100 nm.
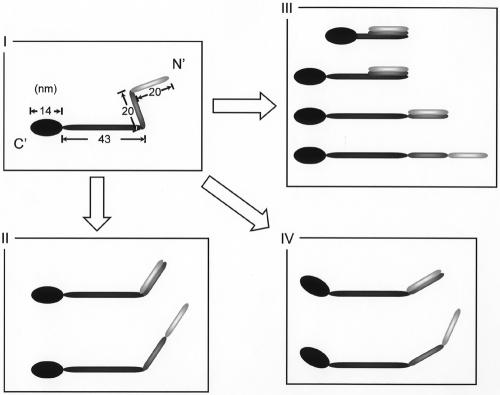
The molecular images were categorized into four types (Fig. 3B), based on the number of bends. Types I and II had two bends and one bend, respectively, while types III and IV had none. Types III and IV featured linear and curved structures, respectively. Some parts of the type IV images looked similar to those of type II but did not show a distinct bend. The most popular type, I, looked like the symbol for an eighth note in music.
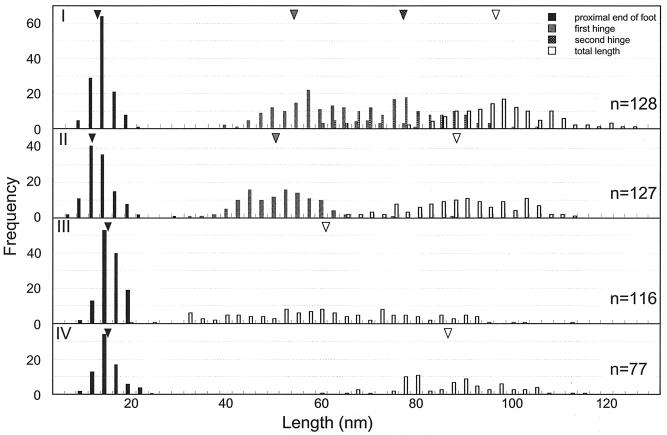
We obtained 494 particle images and analyzed their dimensions and angles, as summarized in Table 1 and Fig. 4 and 5. Types I, II, and III appeared with similar frequencies—29%, 28%, and 26%, respectively—while type IV appeared with lower frequency, 17%. The other 46 particles were not classified into types.
TABLE 1.
Categorization and dimensions of molecular images
| Length (nm)c
|
||||||
|---|---|---|---|---|---|---|
| Typea | Frequencyb (%) | Foot | First rod | Second rod | Third rod | Total |
| I | 29 | 13.6 ± 2.3 | 42.7 ± 6.9 | 20.2 ± 4.1 | 20.0 ± 3.1 | 96.6 ± 9.6 |
| II | 28 | 13.0 ± 3.3 | 37.1 ± 6.9 | 38.6 ± 8.4 | 88.7 ± 12 | |
| III | 26 | 14.8 ± 2.4 | 46.3 ± 18 | 61.1 ± 19 | ||
| IV | 17 | 14.7 ± 2.9 | 71.6 ± 12 | 86.2 ± 11 | ||
The molecular images were categorized into four types, based on the number of bends: types I, II, III, and IV had two, one, no, and no bends, respectively. Types III and IV featured linear and curved structures, respectively.
Classification was done for 494 particle images; 448 images were classified into the four types, while the other 46 were not.
Data are averages ± standard deviations.
FIG. 4.
Positions of features from distal ends of the feet in Gli349 images. The molecule types are indicated on the left in each histogram. Triangles indicate averages.
FIG. 5.
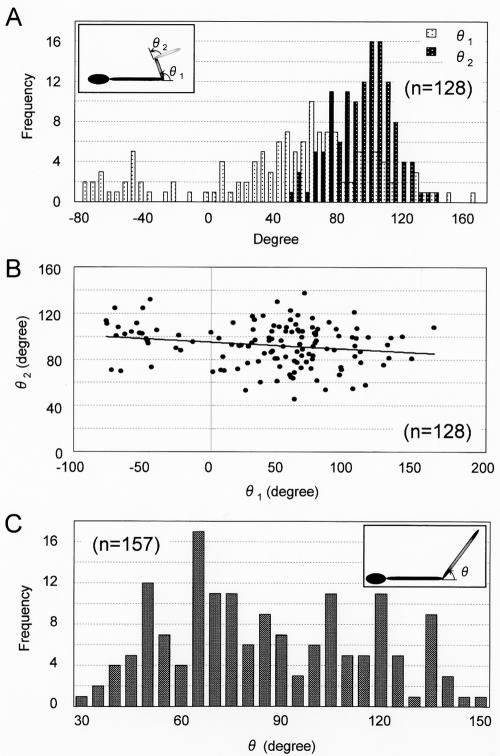
Angles around hinges of type I and II Gli349 images. (A) Distribution of hinge angles θ1 and θ2 in type I images. The inset shows a schematic of the type I image. (B) Relationship between two angles in type I images. The solid line shows the linearized approximation. (C) Distribution of angle θ in type II images. The inset shows a schematic of the type II image.
The type I images consisted of a foot at one end and three rods connected by two hinges, as shown by the illustration in Fig. 3B. The rods and hinges were numbered from the foot side. The angles around the first hinge of the type I images, θ1, ranged from −76 to 161 degrees (Fig. 5A). Multiple Gaussian peak fitting showed that the distribution has moderate preferences at around −52 and 63 degrees (data not shown). On the other hand, the angle around the second hinge, θ2, was fitted by a single Gaussian distribution and nearly constant at 92.3 ± 18.2 degrees. No significant relationship was found between these two angles (Fig. 5B).
Type II images consisted of a foot at one end and two rods connected by a hinge. The features were numbered from the distal end of the foot, as with type I. The angle around the hinge, θ, ranged from 34 to 153 degrees (Fig. 5C). Distinct preferences could not be found even by multiple Gaussian peak fitting (data not shown). The total lengths of type III images, featuring a linear appearance and no hinge, were distributed from 18.9 to 112.2 nm and varied much more widely than the lengths of the other types (Fig. 4). The total lengths of type IV varied less than those of type III, although both types featured an absence of bending.
Gli349 bound by MAb7.
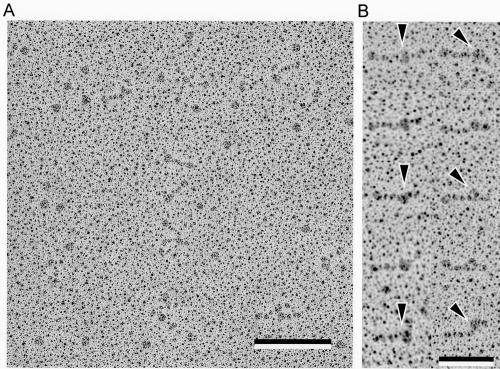
To learn the relationship between the molecular shape of Gli349 and the amino acid sequence, we examined the binding position of a monoclonal antibody, MAb7 (12, 26). MAb7 binds to an epitope localizing in a region comprising amino acids 1193 to 1203 of the 3,183 amino acids in the Gli349 molecule. Gli349 was mixed with MAb7 at the same molar ratio, rotary shadowed, and observed by transmission EM (Fig. 6). Gli349 molecules were found to be bound by a globular particle, MAb7, at similar positions in most molecules. This observation again confirmed the identity between Gli349 and the particle images discussed so far.
FIG. 6.
Rotary-shadowed Gli349 molecules decorated by antibody MAb7. (A) Image 900 nm wide and 900 nm high. Bar, 200 nm. (B) Isolated images of Gli349 molecules coated with MAb7. Foot ends are directed to the left. Arrowheads indicate MAb7 bound to Gli349. Bar, 100 nm.
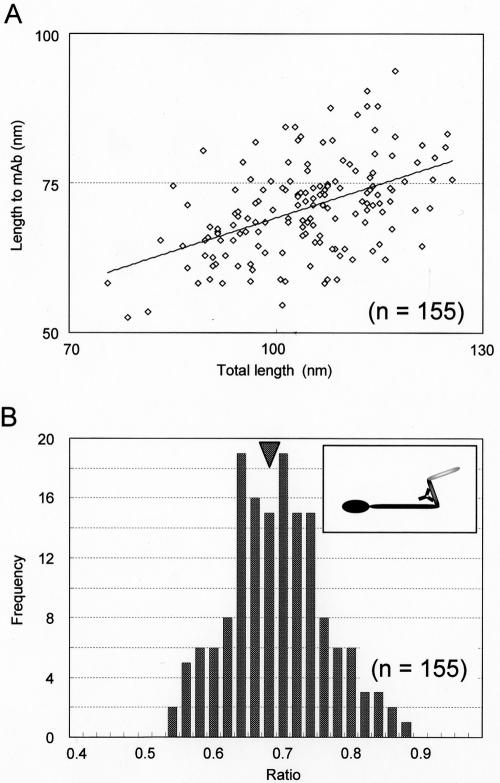
As the foot was easily recognized, we examined the binding positions of MAb7 on the Gli349 molecule (Fig. 7). The average length of the molecule was 104.1 ± 10.0 nm and the length from the foot to MAb7 was 70.8 ± 7.8 nm. The binding position was calculated to be 68.2 ± 7.2% from the foot end. In this analysis, only molecular images longer than 75 nm were used so as to exclude data from folded molecules. The epitope was calculated to be positioned at 0.372 from the N terminus of the total amino acids. Considering this position, the present results show that the foot end corresponds to the C terminus of the amino acid sequence.
FIG. 7.
Positions of MAb7 on molecular images of Gli349. (A) Relationship between positions of MAb7 on Gli349 images and total length. The solid line shows a linearized approximation. (B) Distribution of MAb7 positions on Gli349 images. The central positions of MAb7 are presented as a ratios of the total length of the molecule. The average was 0.682 (n = 155), as indicated by the arrowhead. The inset shows a schematic illustration of a type I molecule coated with MAb7 at the average position.
DISCUSSION
Features of Gli349 suggested from the isolation process.
We tried treating cells with various concentrations of Triton X-100 to optimize the conditions for step 1. Triton X-100 at concentrations higher than 0.075% solubilized Gli349 but the lower ones did not, consistent with the previous observation that treatment of cells with 0.01% Triton X-100 left the gliding machinery capable of reactivation by ATP (27).
In step 2, we found Gli349 in fractions of both 35% and 40% saturation with ammonium sulfate and used the 35% saturation fraction for further studies. To examine the differences between Gli349 proteins from these two fractions, we analyzed the 40% saturation fraction by gel filtration. Gli349 protein was eluted at the same retention time as that of Gli349 from the final fraction presented in Fig. 2 (data not shown). We partially purified Gli349 from the fraction at 40% ammonium sulfate saturation by applying steps 3 and 4 and observed the fraction under EM. The major portion of the images appeared very similar to the images obtained for Gli349 purified from the 35% saturation fraction (data not shown). These observations suggest that the appearance of Gli349 at both 35% and 40% ammonium sulfate saturation was not caused by the heterogeneity of Gli349 by partial association with other proteins, but by the intrinsic character of the molecule. The wide variety of shapes observed by EM may support this explanation.
Shape of the Gli349 molecule.
The images of Gli349 molecules were classified into four types. Assuming that the type I images show most of the molecule's features, the relationships among the types can be portrayed as in Fig. 8, based on the positions of the features and the total lengths (Fig. 4). A foot was observed at one end of each type, showing that all types shared the same structure. The positions of the first hinges in the type I images overlapped for the most part with those of the first hinges in the type II images, suggesting that the first hinges share an identical structure between types I and II.
FIG. 8.
Schematic of relationships among the four types of molecular images. Type I images retain the most features.
The distribution of the total length of type II mostly overlapped the combined positions of the second hinge and the total length in type I images. These results can be explained by the relationship between types I and II presented in Fig. 8. The type III and IV images did not have any hinges. The type III images were linear and varied widely in length from 18.9 to 112.2 nm. These features may suggest that molecules folded in various patterns at the hinges present this type of appearance. The observation that the shorter images tended to be thicker supports this assumption (Fig. 3B). If this assumption is correct, the angle presented as θ2 in the type I images should be 0 or 180 degrees, unlike the preference found in the type I images, which was about 90 degrees.
We did not find any peaks in the distribution of the total length of the type III images, which suggests certain hinges were folded. This failure to find any peaks may have been caused by the variations in molecular length, including occasional fragmentation resulting from the force generated by the spraying and evaporation of the buffer in rotary shadowing. The failure may also have resulted from the rather low resolution of this method. Type IV images, like those of type III, do not have any bends. However, their lengths showed a preferred distribution around 86 nm, like the type II images. These observations suggest that type IV images may be variants of type II, even though their hinges cannot be detected clearly.
Assignment of amino acid sequences to molecular images.
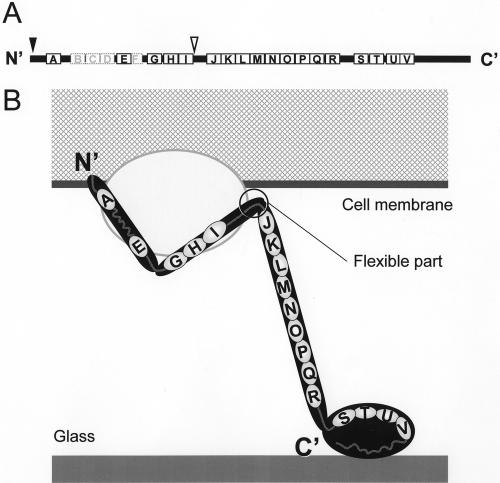
Previously, the amino acid sequence of Gli349 was analyzed with its plausible ortholog, MYPU2110 of M. pulmonis (13). A search of the sequences by using the hidden Markov model revealed that the Gli349 sequence and MYPU2110 have 18 and 22 repeats, respectively, featuring the YXXXXXGF motif and the consensus secondary structure βββα (Fig. 9A). As repeat sequences in general are likely to form rod structures, the amino acid sequence of Gli349 may be assigned to the type I images, based on the following observations. (i) The foot and the third rod correspond to the C-terminal and N-terminal domains, respectively. (ii) A transmembrane segment is predicted near the N terminus. (iii) Although the repeats cannot be detected in the region between repeats A and E using the hidden Markov model in Gli349, three repeats were found in the corresponding region of MYPU2110 of M. pulmonis, suggesting that this region of Gli349 has characteristics similar to those in the repeat regions. (iv) If the repeats tend to form a rod, the nonrepeated parts should be assigned as hinges. (v) The epitope of MAb7 lies in the region comprising the 1,193rd to 1,203rd amino acid on the N-terminal side of the gap region between repeats I and J. We assigned the amino acid sequence on the type I image as presented in Fig. 9B, with which all the observations listed above are consistent.
FIG. 9.
Schematic of the Gli349 molecule. (A) Amino acid sequence. Solid and broken boxes indicate repeats found in Gli349 and positions corresponding to those found only in the ortholog produced by M. pulmonis, MYPU2110, respectively. The names of repeats A to V (13) are indicated on the boxes. The predicted transmembrane segment and epitope of MAb7 are indicated by solid and open triangles, respectively. (B) Amino acid sequence and topology in the cell membrane. The cytoplasm and membrane of a mycoplasmal cell are shown by the meshed area and the thick line, respectively. The glass surface is shown as a gray line. The Gli349 molecule is presented as a rod sticking out of the cell and is supported by other protein components, represented by the gray ellipse. The repeat sequences and gap regions are represented by small ellipses and gray lines, respectively.
Quick-freeze and fracture EM of gliding mycoplasmas revealed that many spike-like structures 50 nm long stick out from the cell membrane and bind to the glass surface with their distal ends (15). The spike is likely to be composed of a Gli349 molecule, based on the following facts (12, 15, 18, 26, 28). (i) The subcellular localization of the spike at the protrusion's base (called the neck) agrees with that of Gli349. (ii) The spikes cannot be found in a mutant in which Gli349 is not localized normally. (iii) A spike's appearance suggests its role in binding to a glass surface during gliding. The rod shape and dimensions of the Gli349 images shown in the present study also support this assumption. The spike-like structure of 50 nm may correspond to 43 nm, which is the length of the first rod of the type I images, or to 56 nm, the sum of the lengths of the foot and the first rod of the images. Considering that the Gli349 molecule has a transmembrane segment near the N terminus, the molecule is likely to be embedded in the gliding apparatus, as presented in Fig. 9B.
Conformational change of the Gli349 molecule in a putative mechanical cycle.
In previous studies, we showed that MAb7 binds to Gli349 of gliding mycoplasmas, reduces the gliding speed, and then displaces the gliding mycoplasmas from the glass surface (12, 26). These observations can be explained by the assumption that the gliding mechanism involves a conformational change of the Gli349 molecule that is responsible for glass binding. To explain the observations obtained so far, we proposed a putative mechanical cycle in which Gli349 moved by or through Gli521 propels the cell by repeating five steps: (i) initial binding to glass, (ii) tight binding to glass, (iii) stroke, (iv) release from glass, and (v) return (14, 24, 26). In the present study, we found that the hinges presented as the first hinges in the type I images are flexible and that the second ones may move among three states represented by angles of 0, 90, and 180 degrees. The variations in molecular images may reflect the conformational changes involved in the mechanical cycle of the gliding mechanism.
MAb7 slows the gliding of M. mobile and finally displaces the cells from the solid surface (12, 26). How does MAb7 cause this? Here, MAb7 was found to bind close to the flexible hinge. Presumably, the binding of MAb7 inhibits the normal conformational changes of Gli349 that are essential for the mechanical cycle, slowing the molecule and causing its detachment from the glass. The Gli349 molecules appeared stiffer when they were bound with MAb7 (Fig. 3 and 6). This observation may support the above assumption.
So far, we have drawn the Gli349 molecule as a rigid leg-like structure (14, 24, 26), because the spike appeared rigid in freeze-fracture EM (15). However, the Gli349 molecules shown here appeared more flexible. Gli349 molecules supported by other components of gliding machinery may form more rigid structures. Alternatively, only molecules at the stage of being rigid in appearance can be visualized by rapid-freeze-and-fracture EM, because a very short moment is fixed and visualized by this fascinating method.
In the present study, we clarified the molecular shape of Gli349, the leg protein involved in the gliding motility of M. mobile, and assigned its amino acid sequence to the image. This information should provide a crucial clue toward elucidating this novel and mysterious biomotility system, although detailed structures of the molecule may be needed to make the story complete.
Acknowledgments
For helpful discussions, we are grateful to Hidetoshi Kono at the Japan Atomic Energy Research Institute and to Naoto Ohtani at Osaka City University.
This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas (genome science) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.M.).
REFERENCES
- 1.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 2.Arata, T. 1998. Electron microscopic observation of monomeric actin attached to a myosin head. J. Struct. Biol. 123:8-16. [DOI] [PubMed] [Google Scholar]
- 3.Bredt, W. 1979. Motility, p. 141-145. In M. F. Barile, S. Razin, J. G. Tully, and R. F. Whitcomb (ed.), The mycoplasmas, vol. 1. Academic Press, New York, N.Y. [Google Scholar]
- 4.Charon, N. W. 2005. Mycoplasma takes a walk. Proc. Natl. Acad. Sci. USA 102:13713-13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez-Patron, C., L. Castellanos-Serra, and P. Rodriguez. 1992. Reverse staining of sodium dodecyl sulfate polyacrylamide gels by imidazole-zinc salts: sensitive detection of unmodified proteins. BioTechniques 12:564-573. [PubMed] [Google Scholar]
- 6.Hiratsuka, Y., M. Miyata, and T. Q. P. Uyeda. 2005. Living microtransporter by uni-directional gliding of Mycoplasma along microtracks. Biochem. Biophys. Res. Commun. 331:318-324. [DOI] [PubMed] [Google Scholar]
- 7.Jaffe, J. D., M. Miyata, and H. C. Berg. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenri, T., S. Seto, A. Horino, Y. Sasaki, T. Sasaki, and M. Miyata. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 11.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 12.Kusumoto, A., S. Seto, J. D. Jaffe, and M. Miyata. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 150:4001-4008. [DOI] [PubMed] [Google Scholar]
- 13.Metsugi, S., A. Uenoyama, J. Adan-Kubo, M. Miyata, K. Yura, H. Kono, and N. Go. 2005. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics 1:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyata, M. 2005. Gliding motility of mycoplasmas—the mechanism cannot be explained by current biology, p. 137-163. In A. Blanchard and G. Browning (ed.), Mycoplasmas: molecular biology, pathogenicity, and strategies for control. Horizon Bioscience, Norfolk, United Kingdom.
- 15.Miyata, M., and J. Petersen. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 186:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 18.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 19.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 169:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence protein and formation of an electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44:431-434. [DOI] [PubMed] [Google Scholar]
- 26.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uenoyama, A., and M. Miyata. 2005. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. USA 102:12754-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uenoyama, A., and M. Miyata. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 187:5578-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]