Abstract
Transcriptional responses to hypoxia are primarily mediated by hypoxia-inducible factors (HIFs), HIF-1α and HIF-2α. The HIF-1α and HIF-2α subunits are structurally similar in their DNA binding and dimerization domains but differ in their transactivation domains, implying they may have unique target genes and require distinct transcriptional cofactors. Our previous results demonstrated that HIF-1α and HIF-2α regulate distinct target genes. Here, we report that HIF-2α is not transcriptionally active in embryonic stem (ES) cells, as well as possible inhibition by a HIF-2α-specific transcriptional repressor. Using DNA microarray analysis of hypoxia-inducible genes in wild-type (WT), Hif-1α−/−, and Hif-2α−/− ES cells, we show that HIF-1α induces a large number of both confirmed and novel hypoxia-inducible genes, while HIF-2α does not activate any of its previously described targets. We further demonstrate that inhibition of HIF-2α function occurs at the level of transcription cofactor recruitment to endogenous target gene promoters. Overexpression of WT and, notably, a DNA-binding-defective HIF-2α mutant restores endogenous HIF-2α protein activity, suggesting that ES cells express a HIF-2α-specific corepressor that can be titrated by overexpressed HIF-2α protein. HIF-2α repression may explain why patients with mutations in the VHL tumor suppressor gene display cancerous lesions in specific tissue types.
Low levels of O2 (hypoxia) are encountered by cells within rapidly growing tissues, such as developing embryos or solid tumors. Most vertebrates respond to this hypoxic stress by activating the expression of a large number of genes involved in glycolysis, angiogenesis, and hematopoiesis (11, 44). This hypoxic transcriptional response is mediated primarily by the hypoxia-inducible transcription factor (HIF), a heterodimer of HIF-α and HIF-β (also called the aryl hydrocarbon receptor nuclear translocator [ARNT]) subunits (48).
HIF activation by hypoxia (≤5% O2) is regulated at the level of α-subunit protein stability in an oxygen-dependent fashion (41). At normoxic O2 levels, HIF-α protein is rapidly degraded due to O2-dependent hydroxylation by prolyl hydroxylase domain-containing proteins and subsequent turnover by a von Hippel-Lindau tumor suppressor protein (pVHL)-dependent degradation pathway. HIF is required for normal embryonic development; ablation of hypoxic responses via targeted deletion of the Hif-α and Arnt genes leads to embryonic lethality (1, 7, 18, 30, 36, 39, 46). On the other hand, an enhanced hypoxic response is a critical component of many cancers (15). For example, the increased glycolysis and angiogenesis observed in most solid tumors is at least partly an effect of elevated levels of HIF activity. Furthermore, constitutive HIF activity resulting from VHL mutations in patients leads to multiple highly vascularized neoplasms in the central nervous system, retina, and kidney (22).
While ARNT is the primary HIF-β subunit, two α subunits, HIF-1α and HIF-2α, participate in the hypoxic responses. HIF-1α is ubiquitously expressed and has been suggested to play a primary role in hypoxic responses. HIF-2α is also widely expressed, but its transcripts are enriched in select cell types, such as vascular endothelial cells, kidney fibroblasts, hepatocytes, glial cells, interstitial cells of the pancreas, epithelial cells of the intestinal lumen, neural crest cell derivatives, and lung type II pneumocytes (19, 47, 52). In contrast to the restricted expression observed in embryonic and adult tissues, HIF-2α is detected in many human tumors, including those associated with VHL disease (renal clear cell carcinomas and hemangiomas) as well as tumors not associated with VHL disease, such as breast, head and neck squamous cell carcinoma, and non-small cell lung cancers (15). Intriguingly, approximately 50% of renal clear cell carcinoma (RCC) cells isolated from VHL patients express HIF-2α but not HIF-1α (32). This expression pattern suggests that HIF-2α plays a tissue-specific role during development and physiology but a broader role in tumorigenesis.
A unique function for HIF-2α in hypoxic cells was not supported by initial studies. For example, both HIF-1α and HIF-2α proteins dimerize with the same ARNT subunit to activate transcription of multiple hypoxia response element (HRE)-dependent reporter genes to similar levels in vitro (47). Furthermore, both HIF-1α and HIF-2α are modified by prolyl hydroxylase domain-containing proteins and the factor inhibiting HIF (FIH) in an O2-dependent manner, giving them similar regulation of protein stability and p300/CBP recruitment (41). The first indication that HIF-1α and HIF-2α play nonredundant roles came from mouse models in which Hif-2α deletion leads to phenotypes distinct from those of Hif-1α−/− embryos (7, 36, 42, 43, 46).
Investigation of the relative roles of HIF-1α and HIF-2α in tumor growth provides additional evidence for distinct functions of the two α subunits. Expression of stabilized HIF-2α but not HIF-1α promotes the growth of xenografts derived from pVHL-reconstituted RCC cells, whereas small interfering RNA (siRNA) knockdown of HIF-2α in pVHL-deficient RCC cells abrogated tumor growth (25, 26, 31). These data suggest that HIF-2α is sufficient and necessary for tumor formation by pVHL-defective RCC cells. Consistent with these findings, we recently observed that teratomas derived from embryonic stem (ES) cells with expression of HIF-2α (but not HIF-1α) from the Hif-1α locus display a substantial growth advantage over wild-types (WT) ES cells (8).
While the distinct function of HIF-2α during development and tumor growth is well established, several important questions remain. For example, what is the molecular mechanism for HIF-2α's unique activity? Does HIF-2α regulate different target genes than HIF-1α? Why do VHL mutations manifest themselves in relatively few tissue-specific tumors despite the fact that pVHL and HIF-2α expression are relatively broad? All mammalian cells, including primary murine ES cells, exhibit hypoxic transcriptional responses. We and others have used ES cells extensively in functional studies of pVHL, HIF-α, and ARNT during hypoxic responses and tumorigenesis (4, 5, 18, 29, 30, 39). WT, Hif-1α−/−, and Hif-2α−/− ES cells provide an attractive system for studying the individual roles of HIF-1α and HIF-2α in response to low O2. We report here that genome-wide DNA microarray analysis of hypoxia-inducible genes in WT, Hif-1α−/−, and Hif-2α−/− ES cells reveals that HIF-2α is not functional in murine ES cells. We demonstrate that expression of HIF-2α mRNA, pVHL regulation of the HIF-2α protein, and formation of nuclear HIF-2α/ARNT complexes are normal in ES cells. Most surprisingly, HIF-2α/ARNT dimer interaction with endogenous HREs is also detected, suggesting that HIF-2α's inability to regulate target gene transcription occurs at the level of transcription cofactor recruitment. HIF-2α overexpression results in detectable HIF-2α activity in Hif-1α−/− ES cells, suggesting ES cells express a HIF-2α-specific corepressor that is titrated by overabundant HIF-2α protein. Importantly, expression of nonfunctional HIF-2α (carrying mutations in the DNA binding domain) but not a similar mutated HIF-1α rescues the function of endogenous HIF-2α protein, presumably by overcoming this repression, while not increasing the levels of functional HIF-2α transcripts. These results provide compelling evidence for the expression of a HIF-2α-specific repressor in ES cells.
MATERIALS AND METHODS
Cell culture.
Hif-1α−/−, Hif-2α−/−, and corresponding WT ES cells were gifts from Peter Carmeliet. Vhl−/− ES cells have been described previously (29). The 786-O WT-8 renal clear cell carcinoma line was a gift from William G. Kaelin (Boston, MA) (17).
RNA preparation, Northern blot analysis, and quantitative RT-PCR.
RNA isolation and Northern blot analysis were performed using standard protocols. Murine DNA fragments generated by reverse transcriptase PCR (RT-PCR) were cloned into the pCR2.1-TOPO vector (Invitrogen) to serve as templates for Northern probes: CaXII (764 to 1544; GenBank accession no. AB005450); Ndrg-1 (1563 to 2916; accession no. gi6679029); Adrp (767 to 1613; accession no. gi191692); Ref (118 to 968; accession no. gi6753085); Ldha (5 to 798; accession no. gi198776); Vegf (129 to 861; accession no. gi202350); Ddit3 (12 to 860; accession no. gi6681148); Adm (413 to 1295; accession no. gi1684819); Upp (6 to 1256; accession no. D44464); Glk (49 to 1181; accession no. gi53931607); Est (24 to 271; accession no. gi12858961); Flot1 (281 to 1592; accession no. AY167925); Rnf19 (1999 to 3451; accession no. BC040769); Slca4 (1563 to 3348; accession no. U75215); Clcn3 (358 to 1759; accession no. gi2599549); Pkp2 (999 to 2346; accession no. gi26340215); Aoc3 (2002 to 3999; accession no. gi3169552); Bing4 (186 to 1876; accession no. gi31982691); Flnb (5000 to 6951; accession no. gi38075550); and Lef1 (1029 to 2399; accession no. gi52887). Mixed primer/probes sets for murine Glut-1, Adm, and 18S rRNA (endogenous control) were used to measure the levels of these transcripts using the Applied Biosystems 7900HT sequence detection system according to the manufacturer's instructions.
cRNA preparation and DNA microarray.
For DNA microarray analysis, triplicate sets of RNA samples were prepared from Hif-1α−/−, Hif-2α−/−, and WT ES cells grown under 1.5% O2 or 21% O2 for 16 h. cDNA and labeled cRNA were generated from these total RNAs. cRNAs were hybridized to Murine Genome Array U74Av2 arrays containing approximately 12,000 distinct mRNA transcripts (Affymetrix). Data analysis was performed using Genechip expression analysis software as described previously (16).
Protein analysis.
Nuclear extracts (NE) were prepared in the presence of protease inhibitors as well as 200 μM deferoxamine (DFX) as described previously (16). Western blot analysis was performed using standard protocols. The following primary antibodies were used for Western blots: anti-HIF-1α monoclonal antibody (Ab) (NB 100-105; Novus Biological, Inc.), anti-HIF-2α polyclonal antibodies (NB 100-122; Novus), anti-ARNT monoclonal antibodies (NB 100-124, Novus), and anti-Flag M2 monoclonal antibodies (F-3165; Sigma). Anti-HIF-α antibodies and protein A-Sepharose CL-4B beads were used to immunoprecipitate HIF-α proteins. Amounts of HIF-α and coprecipitated ARNT protein were assessed using Western blot analysis as described above.
Cloning of HIF-2α mRNA.
Total RNAs isolated from Hif-1α−/−, WT ES, or 786-O WT-8 cells were reverse transcribed using reverse transcriptase and oligo(dT). Primers located at the 5′ or 3′ untranscribed region or different regions of HIF-2α mRNA were used to amplify the full length or different regions of HIF-2α cDNA. The flowing are primers used in this experiment: 5′UTR, 5′-AGACCTTTCACACCTGCCCGGGCGA-3′ (sense); 3′UTR, 5′-GAATATAAATTAGAAGTAATTCAAG-3′ (antisense); F8 (at exon 8), 5′-GCTTCCGAAATCGAGAGAAATAATGG-3′ (antisense); F9 (at exon 11), 5′-GCATGGAGACAGGGACCCTTTG-3′ (antisense); F10 (at exon 8), 5′-CAGAACTTCGATGAACCCTCAGCC-3′ (sense); F11 (at exon 11), 5′-TGTGGCCAGGCCAGCACCCCTC-3′ (sense). PCR products were run on an agarose gel or cloned into the pCR2.1TOPO vector for sequence analysis.
Construction of Hif-1α−/− ES cells expressing a Flag epitope-tagged HIF-1α or HIF-2α protein.
pcDNA3mHIF-1α or HIF-2α plasmids have been described previously (16). The Flag epitope was inserted at the C terminus of HIF-α plasmids in the same reading frame. DNA encoding Flag-tagged HIF-α was transferred into a vector in which the expression of Flag-tagged Hif-α (HIF-αFlag) was under the control of the human elongation factor 1 promoter. DNA-binding-deficient HIF-α mutants (HIF-1αmBHLH and HIF-2α mBHLH) were constructed by changing four conserved basic amino acids in the DNA binding domain of HIF-α subunits to alanines using a PCR-based mutagenesis protocol. The linearized plasmids were electroporated into Hif-1α−/− ES cells, and stable transfectants were selected in the presence of 175 μg/ml hygromycin. Individual clones were picked for Hif-1α−/−/HIF-1αFlag or Hif-1α−/−/HIF-2αFlag, or surviving clones were pooled for Hif-1α−/−/vector, Hif-1α−/−/HIF-1αmBHLH, or Hif-1α−/−/HIF-2αmBHLH.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed using a modified protocol from Emery Bresnick (21). ES cells grown at 1.5% O2 were washed with phosphate-buffered saline containing 100 μm DFX (added to all the solutions). 2 × 107 cells were fixed in 0.4% formaldehyde in phosphate-buffered saline at room temperature for 10 min. Isolated nuclei were lysed, followed by chromatin sonication to sizes between 300 bp and 1,000 bp. Anti-HIF-1α monoclonal Ab (NB 100-105; Novus Biological, Inc.) was used for HIF-1α protein precipitation with mouse immunoglobulin G2b at the same concentration as a control Ab. Anti-HIF-2α pAb (NB 100-122; Novus Biological, Inc.) was used for precipitation of the HIF-2α protein, while rabbit preimmune serum served as a control. After extensive washing, the DNA-protein-Ab complex associated with protein A-Sepharose beads was eluted twice in 100 μl elution buffer. After reversing the cross-linking and purification, DNA from input (1:20 diluted) or immunoprecipitated samples was assayed using regular PCR in the presence of [α-32p]dCTP. The PCR products were separated by acrylamide gel electrophoresis and detected by PhosphorImager analysis. Alternatively, DNA from input or immunoprecipitated samples was quantified by SYBR green real-time PCR. All PCR products were compared to the input amounts to normalize for variations in the input signal that could arise from variable chromatin preparation. The results were plotted as severalfold changes relative to their individual control Ab in individual cells. The primer pairs were pretested to amplify the target genomic DNA in a linear fashion. The following were the primers to detect HRE-containing Vegf and Glut-1 genomic DNA and the Glut-1 promoter region for control in regular Hot-PCR: Glut-1 F, 5′-GGGCTGTGTTACTCACTCTTACTCC-3′; Glut-1 R, 5′-CTCTTCCTGGGTTGTGTTCAAGCTG-3′; VEGF F, 5′-TGTGTGAGAGAGAGAGATCAGGAGG-3′; VEGF R, 5′-GGTGAATGGGATCCTCTGGGAAG-3′; Glut-1 control F, 5′-TAGGTGAGCTGGTTCAATGC-3′; Glut-1 control R, 5′-CTTGCTCCGTGTTCCTGTGC-3′. The following were the primers to detect HRE-containing VEGF and Glut-1 genomic using SYBR-green real-time PCR: Glut-1 F, 5′-ATTTCTAAGGCCCTGGGTCC-3′; Glut-1 R, 5′-CCTGCCTGATGCGTGTCA-3′; VEGF F, 5′-CAGTTGTCTCTCCTTCAGGGCT-3′; VEGF R, 5′-GAAACCCACGTATGCACTGTGTA-3′.
Transient transfection.
The WT HRE-luciferase (Luc) reporter, as well as a mutant HRE-luciferase reporter, was described previously (16). Transient transfection of HEK293 or Hif-1α−/− ES cells with HRE-dependent reporters and/or HIF-α expression plasmids was performed with Lipofectamine reagent with Plus reagent (Invitrogen) according to the manufacturer's instruction.
RESULTS
DNA microarray analysis of hypoxia-inducible genes in WT and Hif-2α−/− ES cells.
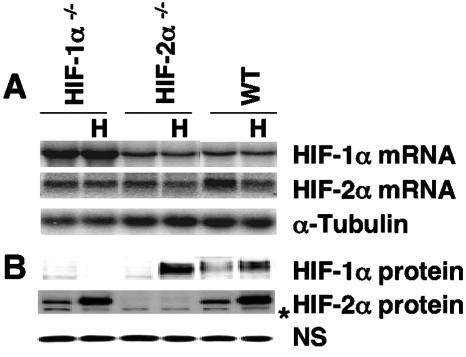
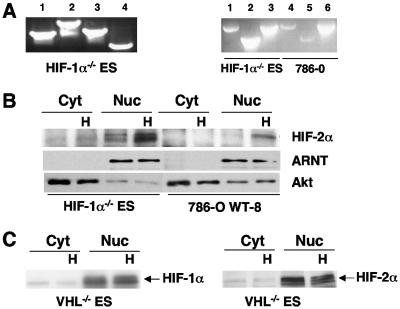
Unlike other mammalian cells, ES cells are highly amenable to the elimination of specific genes via targeted deletions. Hif-1α−/− and Hif-2α−/− ES cells provide a unique opportunity to investigate endogenous HIF-2α or HIF-1α target genes induced by physiological levels of HIF-1α or HIF-2α protein. Therefore, we analyzed Hif-1α−/−, Hif-2α−/−, and corresponding WT ES cells (4, 5). Due to the small number of nucleotides in exon 2, which is deleted in these alleles, the sizes of HIF-1α and HIF-2α mRNAs in mutant ES cells were indistinguishable from those of their WT counterparts (Fig. 1A); however, Northern blot studies clearly showed that ES cells express both HIF-α subunits. Importantly, only the HIF-2α protein was detected in Hif-1α−/− ES cells grown at 1.5% O2 and only the HIF-1α protein in hypoxic Hif-2α−/− ES cells (Fig. 1B), confirming the null mutations of these Hif-1α−/− and Hif-2α−/− ES cells (4, 5).
FIG. 1.
Detection of HIF-1α and HIF-2α mRNA and protein in WT, Hif-1α−/−, and Hif-2α−/− ES cells under normoxia or hypoxia. (A) Northern blot analysis of HIF-1α and HIF-2α mRNA in ES cells grown at 1.5% O2 (H) or 21% O2. (B) Western blot analysis of HIF-α protein in ES cells. The asterisk indicates a nonspecific protein band in the Western blots.
To compare the HIF-1α and HIF-2α target genes, a systematic study of HIF-1α- and HIF-2α-inducible genes was performed on a genome-wide scale. Using an approach we described previously (16), triplicate RNA samples were prepared from WT, Hif-1α−/−, and Hif-2α−/− ES cells cultured under normoxia (21% O2) or hypoxia (1.5% O2) for 16 h. RNA probes generated from these cells were hybridized to the Murine Genome Array U74Av2, containing approximately 12,000 distinct mRNA transcripts (Affymetrix). Comparative analysis of genes expressed at 1.5% O2 versus 21% O2 indicated that hypoxia induced ∼60 distinct genes in the triplicate RNA samples derived from WT ES cells when the threshold indicative of an induced gene was set at 1.4-fold (see Table 1 for some of the induced genes). Hypoxically induced genes included several known HIF target genes, such as vascular endothelial growth factor (Vegf) (7.2-fold in WT ES cells), glucose transporter 1 (Glut-1) (4.1-fold), adipose differentiation-related protein (Adrp) (5.3-fold), adrenomedullin (Adm) (12.2-fold), N-myc downstream regulated 1 (Ndrg-1) (5.3-fold), and most of the glycolytic genes (16). Several novel target genes, such as uridine phosphorylase (Upp) (2.1-fold), adenylate kinase 4 (Ak4) (2.0-fold), annexin A2 (Anxa-2))6.6-fold), and filamin-beta (Flnb) (1.8-fold), were also stimulated (see Table 1, section II, for novel hypoxia-inducible genes confirmed by Northern assays). Multiple known HIF-independent, hypoxia-regulated genes, such as the immediate early response 3 (Ler3) (3.7-fold) (14), DNA-damage-inducible transcripts 3 (Ddit3 or Gadd153) (7.4-fold) (16), and heat shock protein 70 (Hsp70) (1.9-fold) (12) genes, were also induced (Table 1, section III). Surprisingly, the same (95%) genes were also induced at similar levels in Hif-2α−/− cells (Table 1, comparing WT ES cells with Hif-2α−/− ES cells), indicating the dominant role of HIF-1α in the ES cell hypoxic transcriptional response.
TABLE 1.
Hypoxia-inducible genes in WT, Hif-1α−/− and Hif-2α−/− ES cells assessed by microarray analysisa
| Category and gene (ID) | Average fold Induction
|
Description of product | ||
|---|---|---|---|---|
| WT | Hif-1α−/− | Hif-2α−/− | ||
| Section I | ||||
| Gpi1 (L09104) | 1.9 | 1.0 | 2.0 | Glucose phosphate isomerase 1 complex |
| Pgk1 (M15668) | 2.1 | 1.0 | 2.2 | Phosphoglycerate kinase 1 |
| PFK1 (J03928) | 2.3 | 1.0 | 2.2 | Phosphofructokinase, liver, B type |
| SLC2A1 (M22998) | 4.1 | 1.2 | 3.8 | Facilitated glucose transporter, member 1 |
| RTP801 (AK081046) | 2.9 | 1.0 | 3.1 | Regulated in development and DNA damage response |
| ERO1 (AA798624) | 2.5 | 1.0 | 2.5 | ERO1-like (S. cerevisiae) |
| LDH1 (M17516) | 4.2 | 1.5 | 4.0 | Lactate dehydrogenase 1, A chain |
| NDR1 (U60593) | 5.3 | 1.0 | 5.6 | N-myc downstream regulated 1 |
| ADM (U77630) | 12.2 | 1.2 | 10.6 | Adrenomedullin |
| VEGF (M95200) | 7.2 | 1.8 | 6.9 | Vascular endothelial growth factor |
| ADRP (M93275) | 5.3 | 1.0 | 5.1 | Adipose differentiation related protein |
| P4ha1 (U16162) | 4.8 | 1.0 | 3.6 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase, alpha 1 |
| Bnip3 (AF041054) | 4.2 | 1.2 | 4.1 | BCL2/adenovirus E1B 19 kDa-interacting protein 1, NIP3 |
| Stra14 (Y07836) | 3.9 | 1.2 | 3.5 | stimulated by retinoic acid 14 |
| Aldo3 (AW121134) | 3.6 | 1.0 | 3.5 | Aldolase 3, C isoform |
| Ccng2 (U95826) | 4.5 | 1.0 | 3.8 | cyclin G2 |
| Ndr2 (AB033921) | 5.5 | 1.0 | 5.5 | N-myc downstream regulated 2 |
| Stc2 (AF031035) | 3.3 | 1.0 | 3.4 | stanniocalcin 2 |
| Tpi (L31777) | 1.8 | 1.0 | 1.9 | Triosephosphate isomerase |
| Tfr (X57349) | 1.9 | 1.0 | 2.1 | transferrin receptor |
| Igfbp2 (X81580) | 1.8 | 1.0 | 1.8 | insulin-like growth factor binding protein 2 |
| Section II | ||||
| Upp (D44464) | 2.1 | 1.1 | 2.5 | Uridine phosphorylase |
| (AI849432) | 2.6 | 1.0 | 2.8 | EST |
| (AW123751) | 2.0 | 1.0 | 2.1 | EST |
| Anxa2 (M14044) | 6.6 | 1.7 | 6.7 | Annexin A2 |
| PKP2 (AK049481) | 3.2 | 1.0 | 2.9 | Plakophilin 2 |
| Clcn3 (AF029347) | 2.7 | 1.2 | 2.0 | Chloride channel 3 |
| A2M (AI850558) | 8.6 | 1.0 | 7.9 | Alpha-2-macroglobulin |
| AK4 (AB020239) | 2.0 | 1.0 | 1.8 | Adenylate kinase 4 |
| Slca4 (U75215) | 3.7 | 1.0 | 3.9 | Neutral amino acid transporter |
| Flot1 (U90435) | 2.7 | 1.0 | 2.9 | Flotillin 1 |
| Rnf19 (AW12012) | 2.1 | 1.0 | 2.7 | Ring finger protein (C3HC4 type) 19 |
| Aoc3 (AF054831) | 6.3 | 1.2 | 6.1 | Amine oxidase, copper containing 3 |
| Socs3 (U88328) | 1.9 | 1.0 | 1.8 | Suppressor of cytokine signaling 3 |
| Lef1 (X58636) | 2.9 | 1.0 | 3.0 | Lymphoid enhancer binding factor 1 |
| GLK (AB027012) | 1.8 | 1.0 | 1.8 | Galactokinase |
| Flnb (AI591702) | 1.8 | 1.0 | 1.8 | Filamin, beta |
| Section III | ||||
| Ler3 (X67644) | 3.7 | 3.5 | 4.1 | Immediate early response 3 |
| Ddit3 (X67083) | 7.4 | 6.9 | 7.3 | DNA-damage inducible transcript 3 |
| Hsp70-4 (U08215) | 1.9 | 1.6 | 1.9 | Heat shock protein, 70 kDa 4 |
Genes induced in triplicate sets of hypoxic mRNAs obtained from WT, Hif-1α−/−, and Hif-2α−/− ES cells as determined by DNA microarray assays. Target gene induction represents hypoxic levels relative to normoxic levels of expression. Section I represents select known hypoxia-inducible genes. Section II contains novel hypoxia-inducible genes, confirmed by Northern blot analysis. Section III includes known HIF-independent hypoxia-inducible genes. ID, GenBank accession no.
Hif-1α−/− ES cells exhibit no hypoxic gene induction.
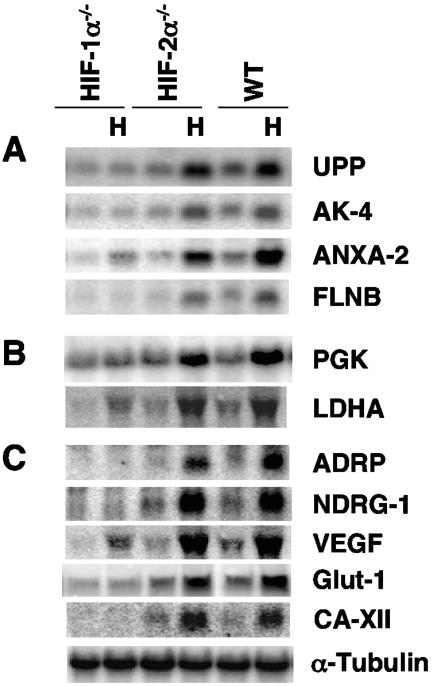
Most of the genes stimulated by hypoxia in WT and Hif-2α−/− ES cells were not induced in Hif-1α−/− ES cells (Table 1). Only about eight genes were induced in Hif-1α−/− ES cells in the triplicate RNAs according to DNA microarray analysis. The three most highly induced genes, Ler3, Ddit3, and Hsp70, exhibited levels of hypoxic induction similar to those observed in the WT and Hif-2α−/− ES cells (Table 1, section III). These genes are known to be HIF-independent, stress-inducible genes (12, 14, 16). The next three genes, Vegf (1.8-fold), Anxa-2 (1.7-fold), and lactate dehydrogenase A (Ldha) (1.5-fold), exhibited lower levels of induction in the Hif-1α−/− ES cells than in the WT and Hif-2α−/− ES cells (Table 1). Two additional genes induced in Hif-1α−/− ES cells, CD68 antigen (CD68) (1.4-fold) and histone gene complex 1 (Hist1) (1.4-fold), exhibited even lower stimulation. Northern blot analysis of 16 novel hypoxia-inducible genes confirmed that 15 of them (Table 1, section II) were indeed stimulated in both WT and Hif-2α−/− ES cells but not in Hif-1α−/− ES cells (Fig. 2A; also data not shown). Of note, Anxa2 was minimally induced in Hif-1α−/− ES cells. The induction of CD68 and Hist1 was not confirmed by Northern assays (data not shown). In contrast, analysis of several known HIF-1α unique genes, such as phosphoglycerate kinase 1 (Pgk-1) and Ldha (Fig. 2B), and HIF-1α/HIF-2α common target genes (Adrp, Ndrg-1, Vegf, Glut-1, Ca12, and Adm) (Fig. 2C and data not shown) confirmed the DNA microarray analysis. Interestingly, lower levels of induction of Vegf and Ldha were detected in Hif-1α−/− ES cells.
FIG. 2.
Northern blot analysis of induced genes observed in the DNA microarray confirms that all of these genes are induced in hypoxic WT and Hif-2α−/− ES cells. While most of these genes lost hypoxic induction in Hif-1α−/− ES cells, the annexin A2 (Anxa-2), lactate dehydrogenase A (Ldha), and vascular endothelial growth factor (Vegf) genes are induced in Hif-1α−/− ES cells, albeit at lower levels than in WT and Hif-2α−/− ES cells. (A) Novel target genes: uridine phosphorylase (Upp), adenylate kinase 4 (Ak4), Anxa-2, and filamin-beta (Flnb). (B) Two known HIF-1α-specific genes: phosphoglycerate kinase 1 (Pgk-1) and Ldha. (C) Known HIF-2α/HIF-1α common target genes: adipose differentiation-related protein (Adrp), N-myc downstream regulated 1 (Ndrg-1), Vegf, glucose transporter 1 (Glut-1), and carbonic anhydrase 12 (CaXII).
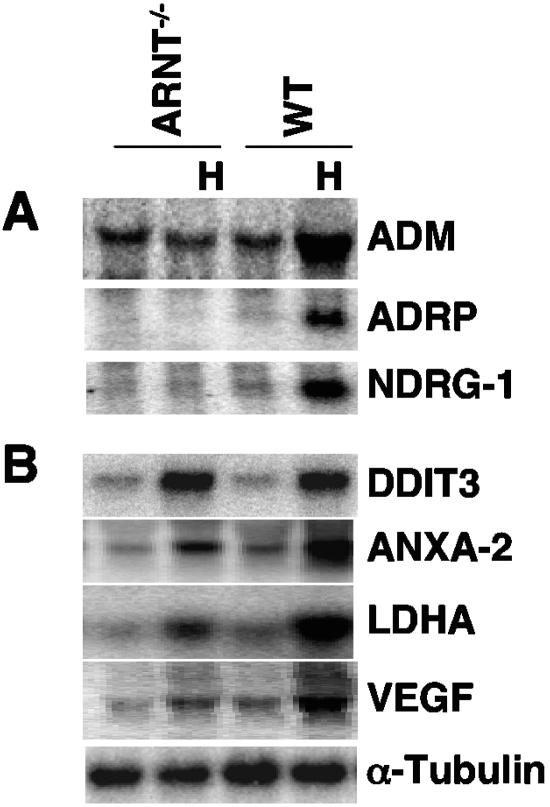
The reduced induction of Anxa2, Vegf, and Ldha in Hif-1α−/− ES cells showed that they were clearly regulated by HIF-1α. Furthermore, induction of these three genes in Hif-1α−/− ES cells also suggested that HIF-2α regulates their expression. Alternatively, induction of these genes in Hif-1α−/− ES cells may be HIF independent and regulated by other stress-induced transcription factors or hypoxic mRNA stabilization, as reported for Vegf (45). To distinguish between these two possibilities, we performed Northern blot analysis of each gene in Arnt−/− ES cells. Loss of ARNT results in the complete abolition of HIF-dependent hypoxic gene induction in ES cells, since ES cell does not express ARNT-2 (30). As expected, hypoxic stimulation of several HIF-1α/HIF-2α common target genes (Adm, Adrp, and Ndrg-1) was eliminated in Arnt−/− ES cells (Fig. 3A). Consistent with our previous results (16), hypoxic upregulation of Ddit-3 was not dependent on Arnt status, since it was similarly induced in both WT and Arnt−/− ES cells (Fig. 3B). Importantly, hypoxic induction of Anxa-2, Ldha, and Vegf was reduced in Arnt−/− ES cells (Fig. 3B), indicating that they are HIF transcriptional targets. This was consistent with the above data showing that HIF-1α elimination decreased their hypoxic induction (Fig. 2 and Table 1). Low-level induction of these genes in Arnt−/− ES cells suggests that hypoxic stimulation observed in Hif-1α−/− ES cells resulted from HIF-independent mechanisms, such as changes in mRNA stability. Thus, the results from the DNA microarray analysis of 12,000 mRNAs in ES cells and Northern blot analysis of a selected group of genes demonstrated that Hif-1α−/− ES cells exhibited no HIF-mediated target gene induction.
FIG. 3.
Northern blot analysis of hypoxia-inducible genes in Arnt−/− ES cells. (A) Hypoxic induction of Adm, Adrp, and Ndrg-1 is abolished in Arnt−/− ES cells. (B) Hypoxic induction of DNA-damage-inducible transcript 3 (Ddit3 or Gadd153) is HIF independent, while hypoxic induction of Anxa-2, Ldha, and Vegf in WT ES cells constitutes the combined effects of HIF and HIF-independent regulation.
HIF-2α is inactive in Hif-1α−/− ES cells.
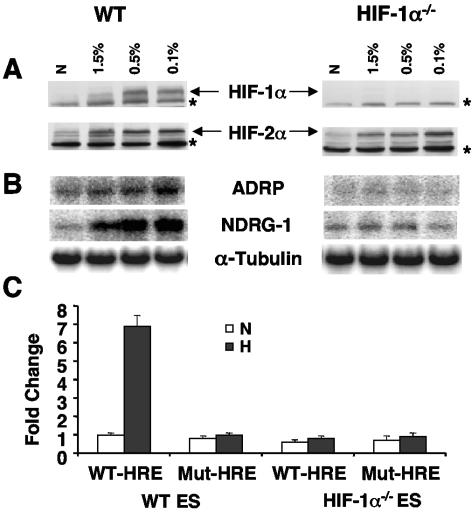
We showed previously that severe hypoxia (0.01%) induces HIF target gene expression to higher levels in pVHL-reconstituted 786-O WT-8 renal cells than 1.5% O2 (16). We therefore tested whether treatment of Hif-1α−/− ES cells with lower levels of O2 (0.5% and 0.1% O2) induced HIF-2α target gene expression in these cells. As expected, WT ES cells exhibited elevated levels of HIF-1α/HIF-2α common target gene expression under severe hypoxia (Fig. 4B, left), presumably due to higher levels of HIF-α protein expression under severe hypoxia (Fig. 4A, left). In contrast, no HIF target gene induction was observed in Hif-1α−/− ES cells, although elevated levels of HIF-2α protein were present (Fig. 4B and 4A, right). Furthermore, several hypoxia mimetics, including DFX and cobalt chloride, also failed to induce several HIF-2α target genes in Hif-1α−/− ES cells, although the HIF-2α protein was stabilized under these treatments (data not shown). In addition, 16 h of hypoxia failed to enhance the expression of transfected WT HRE-Luc reporter constructs in Hif-1α−/− ES cells, while the WT HRE-Luc reporter exhibited a sevenfold hypoxic induction in WT ES cells (Fig. 4C). Therefore, analysis of endogenous HIF target genes by genome-wide DNA microarray and more-sensitive HRE-mediated reporter assays demonstrated that endogenous HIF-2α is not functional in murine ES cells.
FIG. 4.
Hif-1α−/− ES cells exhibit no HIF-2α activity under severe hypoxia or by activating HRE reporters. (A) Severe hypoxia increases the levels of HIF-1α and HIF-2α protein in WT ES cells (left panel) and HIF-2α protein levels in the Hif-1α−/− ES cells (right panel). Nonspecific proteins in HIF-1α or HIF-2α Western blots labeled by asterisks serve as loading controls. (B) Severe hypoxia increases HIF-α common target gene expression in WT ES cells (left panel) but not in Hif-1α−/− ES cells (right panel). (C) Hypoxic treatment of Hif-1α−/− ES cells fails to enhance the transfected WT HRE reporter gene, while WT ES cells exhibit hypoxic induction of transfected WT-HRE reporter construct. Results are plotted as changes relative to a mutant (mt-HRE) reporter under normoxia.
Hif-1α−/− ES cells express full-length HIF-2α mRNA and exhibit normal pVHL regulation of the HIF-2α protein.
The fact that HIF-2α is functional in both 786-O WT-8 and HEK293 Tet-on HIF-2DPA cells that express HIF-2α only (16) argues strongly against the idea that HIF-1α is required for HIF-2α activity. For HIF-2α to activate its target genes, a number of events must occur. Therefore, we investigated the expression of HIF-2α transcripts, hypoxic stabilization of the HIF-2α protein, subcellular location of stabilized HIF-2α protein, binding of HIF-2α with its partner ARNT, and binding of HIF-2α/ARNT to HREs of HIF-2α target genes in Hif-1α−/− ES cells.
Multiple splicing variants of HIF-1α (but not HIF-2α) have been reported (13, 51). To investigate whether Hif-1α−/− ES cells expressed a cell-specific HIF-2α isoform missing an important exon or a mixture of full-length with inhibitory variants, we performed RT-PCR cloning and sequence analysis of HIF-2α mRNA from Hif-1α−/− and WT ES cells. A single band exhibiting the expected size was generated using HIF-2α primers designed to amplify segments from the 5′ UTR to amino acid (aa) 418 (F8) (Fig. 5A, left, lane 1) or to aa 619 (F9) (lane 2) and aa 418 (F10) (lane 3) or aa 619 (F11) (lane 4) to the 3′ UTR. Furthermore, identical sizes of HIF-2α cDNAs were generated from murine Hif-1α−/− ES cells and the HIF-2α-functional human 786-O cell line using murine primers to amplify the full length (Fig. 4A, right, lanes 1 and 4), 5′UTR to aa 619 (lanes 2 and 5), and to aa 851 (lane 3 and 6). Importantly, sequence analysis of several cloned HIF-2α fragments, including six full-length mHIF-2α clones from Hif-1α−/− ES cells (using primers located at the 5′UTR and 3′UTR), determined that ES cells expressed full-length HIF-2α mRNA with no splicing variants.
FIG. 5.
Hif-1α−/− ES cells express a full-length HIF-2α mRNA whose protein product is normally regulated by pVHL. (A) RT-PCR cloning of HIF-2α cDNA from Hif-1α−/− ES cells using primers to amplify the different regions of HIF-2α (left panel); cloning of full-length HIF-2α or HIF-2α fragments from Hif-1α−/− ES cells versus 786-O WT-8 cells exhibiting functional HIF-2α (right panel). (B) HIF-2α protein is primarily detected in the nucleus of hypoxic Hif-1α−/− ES cells (left panel), as observed in 786-O WT-8 (right panel). ARNT (a nuclear protein) and Akt served as loading controls for the quality of cellular fractions. Antibodies to HIF-2α, ARNT, and Akt detect both murine and human proteins. (C) Loss of pVHL in Vhl−/− ES cells results in normoxic stabilization of nuclear HIF-1α (left panel) and HIF-2α (right panel) protein.
A recent paper has suggested that HIF-2α is not functional in murine embryonic fibroblasts (MEFs) because HIF-2α is sequestered in the cytoplasm and immune to pVHL regulation (35). Western blot analysis of the HIF-2α protein (using antibody recognizing both the murine and human HIF-2α proteins) on cytoplasmic and nuclear fractions from ES cells showed that HIF-2α accumulated in hypoxic Hif-1α−/− ES cells and was localized to the nucleus, as observed for HIF-2α in 786-O WT-8 cells (Fig. 5B). Detection of the nuclear protein ARNT only in nuclear fractions and of Akt primarily in cytoplasmic fractions validated the quality of these extracts (Fig. 5B). In addition, HIF-2α protein accumulation was regulated by pVHL in ES cells, since Vhl deletion stabilized the HIF-2α protein under normoxia and both the HIF-1α and HIF-2α proteins translocated to the nucleus (Fig. 5C). Therefore, cytoplasmic trapping of HIF-2α, as described previously for MEFs (35), is not involved in HIF-2α inhibition in ES cells.
HIF-2α protein forms a complex with ARNT and binds to HREs of target genes in Hif-1α−/− ES cells.
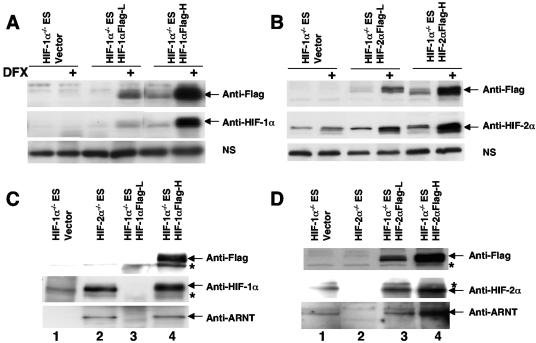
It is formally possible that HIF-2α and ARNT do not form dimers in ES cells. Anti-HIF-2α antibodies recognize mouse HIF-2α protein but also several nonspecific cellular proteins. To provide an additional positive control for HIF-α detection (especially in immunoprecipitation experiments [IPs]), we generated cell lines in which Hif-1α−/− ES cells were stably transfected with C-terminal Flag epitope-tagged full-length HIF-1α (Fig. 6A) or HIF-2α cDNA (Fig. 6B) under the control of the human elongation factor 1 promoter. Addition of the Flag tag did not change the subcellular location and function of HIF-α proteins, since Flag-tagged HIF-α proteins were detected only in nuclear fractions of hypoxic cells and activated cotransfected HRE-mediated reporters in transient transfections (data not shown). As shown in Fig. 6A, anti-HIF-1α antibodies detected HIF-1α protein in nuclear fractions isolated from cells stably transfected with HIF-1αFlag cDNA, not in parental Hif-1α−/− ES cells. Anti-Flag Western blot analysis confirmed the anti-HIF-1α Western blot result (Fig. 6A). Hif-1α−/− ES cells expressed the endogenous HIF-2α protein, and transfection of HIF-2α cDNA into these cells increased levels of the HIF-2α protein as detected by anti-HIF-2α antibody (Fig. 6B). The identity of the HIF-2α band was confirmed by anti-Flag Western blot analysis (Fig. 6B).
FIG. 6.
Endogenous HIF-2α forms a complex with ARNT in Hif-1α−/− ES cells. Establishment of Hif-1α−/− ES cells stably expressed low (L) or high (H) levels of Flag epitope-tagged HIF-1α protein (A) or HIF-2α protein (B) as assessed by Western blot analysis using anti-Flag or anti-HIF-α antibodies in the nuclear extracts derived from deferoxamine (DFX)-treated ES cells. Nonspecific protein bands (labeled NS) in the HIF-α Western blots served as loading controls. (C) Anti-HIF-1α antibody was used to immunoprecipitate HIF-1α protein in the nuclear extracts derived from hypoxia (1.5% O2)-treated ES cells. The amount of precipitated HIF-1α and coprecipitated ARNT protein was assessed using Western blot analysis. (D) Anti-HIF-2α antibody was used to immunoprecipitate HIF-2α in the nuclear extracts derived from hypoxia (1.5% O2)-treated ES cells. ARNT protein is coprecipitated with the HIF-2α protein in Hif-1α−/− ES cells (lane 1). Nonspecific protein bands (labeled by an asterisk) in anti-Flag Western blots (for panels C and D) exhibited similar intensities in all lanes.
We then tested the formation of HIF-1α/ARNT and HIF-2α/ARNT complexes in ES cells. We were particularly interested in the formation of endogenous HIF-2α/ARNT dimers, since HIF-2α in Hif-1α−/− ES cells was not functional. Hif-1α−/− ES/HIF-1αFlag-H and Hif-1α−/− ES/HIF-2αFlag-H (high) clones served as positive controls in IPs, since these cells exhibited HIF-α-mediated target gene expression (see Fig. 8) and were expected to exhibit HIF-αFlag/ARNT complexes. In addition, the identities of HIF protein bands could be confirmed by independent anti-Flag Western blotting. NEs were prepared from ES cells grown at 1.5% O2 for 8 h, and equivalent NE quantities were used for precipitation by anti-HIF-1α or anti-HIF-2α antibody. The precipitated HIF-α and coprecipitated ARNT proteins were assessed using identical volumes of IP material. Anti-HIF-1α antibodies immunoprecipitated endogenous HIF-1α protein in Hif-2α−/− ES cells (Fig. 6C, lane 2) and transfected HIF-1αFlag protein in Hif-1α−/−/HIF-1αFlag-H cells (Fig. 6C, lane 4), as detected by anti-HIF-1α and confirmed by anti-Flag Western blot analysis (Fig. 6C). Coprecipitation of ARNT with endogenous HIF-1α or transfected Flag-tagged HIF-1α indicated that the HIF-1α/ARNT dimers were present in ES cells (Fig. 6C, lanes 2 and 4), consistent with functional data that HIF-1α induces its target genes in Hif-2α−/− ES cells (Fig. 2 and Table 1) or in Hif-1α−/−/HIF-1αFlag-H cells (see Fig. 8C). Anti-HIF-2α antibody precipitated endogenous HIF-2α protein (Fig. 6D, lane 1) as well as transfected HIF-2αFlag protein (Fig. 6D, lanes 3 and 4), as detected by anti-HIF-2α and confirmed by anti-Flag antibodies. Importantly, coprecipitated ARNT protein was detected only in IPs where HIF-2α protein was detected (Fig. 6D, lanes 1, 3, and 4). In addition, the amount of coprecipitated ARNT correlated well with the amount of the HIF-2α protein precipitated. The detection of ARNT protein in the HIF-2α IP from Hif-1α−/− ES cells demonstrated that endogenous HIF-2α formed a complex with endogenous ARNT protein in ES cells.
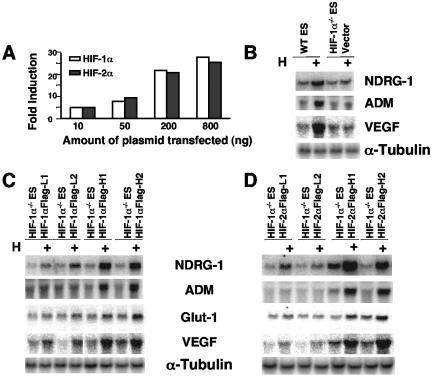
FIG. 8.
Increased expression of WT HIF-2α in Hif-1α−/− ES cells renders HIF-2α functional in ES cells. (A) Transient transfection of either HIF-1α or HIF-2α expression plasmid induces a cotransfected HRE-dependent reporter gene in Hif-1α−/− ES cells in a dose-dependent manner. Results are plotted as changes relative to a vector control under normoxia. (B) HIF-1α/HIF-2α common target genes are induced in hypoxic WT but not in Hif-1α−/− ES cells transfected with vector expressing the Flag peptide as analyzed by Northern blotting. (C) WT levels of HIF-1α expression (H1 and H2 have similar levels of HIF-1α detected in WT ES cells) restore HIF target gene induction, while 20% of WT levels of HIF-1α expression (L1 and L2) restore target gene induction partially. (D) High levels of HIF-2α protein (H1 and H2 express the HIF-2α protein at sevenfold higher levels than that detected in Hif-1α−/− ES cells) expression are required for HIF-2α activity, while low levels of HIF-2α expression (L1 and L2 express threefold levels relative to that of WT ES cells) restore target gene induction marginally.
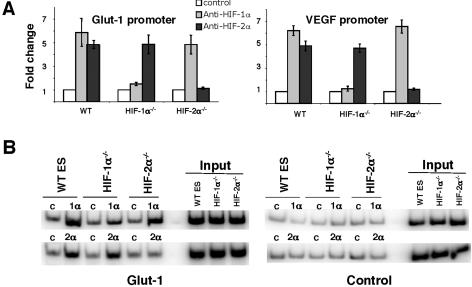
Redox effector factor 1 (Ref-1) appears to be critical for HIF-2α DNA binding activity (27). Failure to express Ref-1 would prevent HIF-2α (but not HIF-1α) from binding to target gene HREs in ES cells and would consequently inactivate HIF-2α. We determined that levels of Ref-1 (mRNA and protein) in ES cells were similar to those in HEK293 and 786-O cells, two cell types known to harbor functional HIF-2α (data not shown), suggesting that Ref-1 is not involved in the inhibition of HIF-2α activity in ES cells. To directly test whether endogenous HIF-2α binds to the HREs of endogenous ES cell target genes, we performed ChIP on WT, Hif-1α−/−, and Hif-2α−/− ES cells. Glut-1 and Vegf are HIF-1α/HIF-2α common target genes with well-characterized HREs (see details in Discussion). Anti-HIF-1α or -HIF-2α antibodies were used to precipitate the HIF-1α or HIF-2α protein, and the amount of coprecipitated HRE-containing genomic DNA fragments from Glut-1 and Vegf promoters was assessed by quantitative SYBR green-based real-time PCR (Fig. 7A), as well as regular PCR using 32P-labeled dCTP (Fig. 7B). While isotype-matched control antibodies generated similar signals from WT, Hif-1α−/− ES, or Hif-2α−/− ES cells, anti-HIF-1α antibody enriched HRE-containing Glut-1 and Vegf genomic DNA fragments in WT and Hif-2α−/− ES cells but not in HIF-1α-deficient Hif-1α−/− ES cells (Fig. 7A), suggesting that HIF-1α antibodies specifically pull down HIF-1α-associated genomic DNA fragments and HIF-1α protein interacted with the HREs of its two target genes Glut-1 and Vegf in hypoxia-treated cells. This was consistent with the fact that the HIF-1α protein stimulated Glut-1 and Vegf gene expression in WT and Hif-2α−/− ES cells (Fig. 2C and Table 1). IP with anti-HIF-2α antibody generated a stronger signal using lysates prepared from HIF-2α-expressing hypoxic WT and Hif-1α−/− ES cells and not from Hif-2α−/− ES cells (Fig. 7A). In agreement with quantitative SYBR Green-based real-time PCR, detection of coprecipitated Glut-1 HREs using primers covering the whole Glut-1 HRE also indicated that HIF-1α or HIF-2α bound to the Glut-1 promoter in its expressing cells only (Fig. 7B, left). In contrast, a flat signal was detected in all cell types using primers located at the murine Glut-1 promoter (about 2.5 kB downstream of Glut-1 HREs) (Fig. 7B, right), indicating optimum sonication of genomic DNA and specificity of ChIP. The chromatin IP results indicated that endogenous HIF-2α, like HIF-1α, bound to HREs of its target genes in ES cells. Moreover, these data argue that HIF-2α inhibition in ES cells occurs at the stage of transcriptional cofactor recruitment at endogenous promoters.
FIG. 7.
The endogenous HIF-2α protein occupies the HREs of Glut-1 and Vegf in Hif-1α−/− ES cells in vivo, as shown by chromatin IP. (A) Anti-HIF-1α or anti-HIF-2α or control antibodies (mouse immunoglobulin G2b for anti-HIF-1α and rabbit preimmune serum for anti-HIF-2α) were used to precipitate HIF-α protein in cross-linked hypoxic ES cells. Coprecipitated DNA fragments were detected using SYBR green real-time PCR by primers specific for HREs of two HIF-1α and HIF-2α common target genes, Glut-1 and Vegf. Results were the averages for three independent experiments and were plotted as changes relative to their individual control antibodies. (B) ChIP assays as detected by regular PCR labeled with [32P]dCTP using primers spanning the HRE of the Glut-1 enhancer to assess HIF binding (Glut-1, left panel) or primers locating at 2.5 kb downstream of Glut-1 HRE-containing enhancer to assess experimental specificity (Control, right panel). A representative assay from three independent experiments is shown. “1α”, “2α,” and “c” indicate anti-HIF-1α, anti-HIF-2α, or control antibodies for immunoprecipitation.
Overexpression of HIF-2α in Hif-1α−/− ES renders HIF-2α active.
We hypothesized that HIF-2α transcriptional activity either requires an activator not expressed in ES cells or is inhibited by a repressor present in ES cells. If HIF-2α were repressed, then overexpression of the HIF-2α protein might saturate repressor binding and allow repressor-free HIF-2α to activate its target gene expression in ES cells. Transient transfection of 10 ng HIF-1α or HIF-2α expression plasmid into 35-mm dishes allowed detection of the HIF-α protein (see Fig. S1 in the supplemental material) and induced expression of a cotransfected WT HRE-Luc reporter gene in Hif-1α−/− ES cells, demonstrating that HIF-2α can function in Hif-1α−/− ES cells if sufficiently overexpressed (Fig. 8A). Interestingly, HIF-2α induction of the reporter gene was very similar to that of HIF-1α, even at low plasmid doses. This suggested that even 10 ng HIF-2α expression plasmid resulted in HIF-2α overexpression, making transient transfection unsuitable for analysis of a possible HIF-2α repression mechanism in ES cells. To investigate whether relative HIF-2α overexpression would rescue endogenous HIF-2α activity in ES cells, we used our established Hif-1α−/− ES cell lines stably transfected with different levels of Flag-tagged HIF-2α or HIF-1α cDNAs as described in the legend to Fig. 6. These cells also allow us to investigate HIF-α activity at endogenous HIF target gene promoters. As shown in Fig. 8B, induction of several HIF-1α/HIF-2α common target genes was observed in the WT ES cells but not in the Hif-1α−/−/vector ES cells. As expected, reintroduction of HIF-1α to levels similar to that observed in WT ES cells (two Hif-1α−/−/HIF-1αFlag-High clones) restored hypoxic gene regulation completely (Fig. 8C), while the two Hif-1α−/−/HIF-1αFlag-Low clones (at 20% of levels seen in WT ES cells) restored target gene induction only partially. Hypoxic induction of HIF-2α target genes was achieved in two Hif-1α−/−/HIF-2αFlag-High clones (Fig. 8D) that expressed the HIF-2α protein at sevenfold higher levels than endogenous HIF-2α (Fig. 6B and D), while limited activation of Ndrg-1 (but not Adm, Glut-1, and Vegf) was observed in the two Hif-1α−/−/HIF-2αFlag-Low clones (Fig. 8D) that expressed the HIF-2α protein at threefold-higher levels than endogenous HIF-2α (Fig. 6B and D). While Hif-1α−/−/HIF-1αFlag-High clones exhibited hypoxic induction of HIF-1α unique genes (Pgk and Ldha), hypoxic stimulation of these genes was not observed in the Hif-1α−/−/HIF-2αFlag-High clones (data not shown). Therefore, restoration of endogenous target gene induction was the direct function of transfected HIF-α cDNA, and not the stabilization of endogenous HIF-2α protein. These results are consistent with a model in which overexpressed HIF-2α restores the function of HIF-2α by titrating repressor(s) that negatively regulate HIF-2α function. To further test this model, we constructed mutated HIF-α constructs (HIF-1αmBHLH and HIF-2αmBHLH) in which several highly conserved basic amino acids in HIF-α DNA binding domains were mutated to alanines (HIF-2α, -R24C25R26R27S28K29-to -A24C25A26A27S28A29-). We hypothesized that such constructs would bind to HIF-interacting factors normally but would not directly participate in target gene transcription. A similar HIF-2α mutant was previously shown to lack both DNA binding and target gene induction capabilities (25). In contrast to the case with WT HIF-1α and HIF-2α plasmids, transient transfection of the HIF-2α and HIF-1α mutants failed to induce the activity of a cotransfected WT-HRE-Luc reporter in HEK 293 cells under normoxia (Fig. 9A). Of note, mutated HIF-αmBLHL protein was detected only in the nuclear fraction of the transfected cells (data not shown). Furthermore, hypoxia treatment (stabilizing endogenous HIF-2α protein) did not induce HRE reporter gene activity in Hif-1α−/− cells transfected with either HIF-1αmBHLH or HIF-2αmBHLH, indicating that HIF-2αmBHLH expression did not rescue endogenous HIF-2α activity (data not shown). To eliminate the overwhelming expression characteristic of transient transfections, a Hif-1α−/− ES cell population stably transfected with HIF-αmBHLH was established. Interestingly, hypoxic treatment increased Glut-1 and Adm (HIF-1α/HIF-2α common target genes) expression in the Hif-1α−/− ES cells stably transfected with the HIF-2α mutant (2.2-fold for Glut-1 and 4.1-fold for Adm) but not in cells transfected with HIF-1αmBHLH or vector (Fig. 9B). Since the HIF-2α mutant cannot bind to DNA, this induction could only come from the endogenous HIF-2α protein. These results argue that the HIF-2α mutant enhanced activity of endogenous HIF-2α by titrating away a HIF-2α repressor, and this repressor is specific for HIF-2α. These data strongly suggest that a HIF-2α-specific repressor negatively regulates endogenous chromatin-associated HIF-2α activity in ES cells.
FIG. 9.
Expression of a DNA-binding-defective HIF-2α mutant (HIF-2αmBHLH) but not a similarly mutated HIF-1α construct rescues the function of endogenous HIF-2α in the Hif-1α−/− ES cells. (A) Both HIF-2αmBHLH and HIF-1αmBHLH mutants do not induce a cotransfected WT-HRE reporter in HEK293 cells under normoxia, while the same amount of WT HIF-α plasmids induces WT-HRE reporter. Results were plotted as change relative to that of empty vector under normoxia. (B) Hypoxia induces Glut-1 and ADM (HIF-1α and HIF-2α common genes) expression in WT and Hif-1α−/−/HIF-2αmBHLH ES cells as assessed by real-time PCR but not in Hif-1α−/−/HIF-1αmBHLH or Hif-1α−/−/vector ES cells. Results are the averages for three independent experiments and are plotted as changes relative to that in Hif-1α−/−/vector cells under normoxia.
DISCUSSION
Functional studies of HIF-1α and HIF-2α are complicated by the fact that most cell lines express both HIF-1α and HIF-2α. Elimination of HIF-1α expression in Hif-1α−/− ES cells and HIF-2α expression in Hif-2α−/− ES cells provides us with a good opportunity to investigate the relative contributions of HIF-1α and HIF-2α to hypoxic responses. Results from previous studies indicate that HIF-2α in Hif-1α−/− ES cells does not regulate a number of glycolytic genes (4, 5, 18, 39). Recently, we and others found that glycolytic genes are preferentially regulated by HIF-1α (16, 49). Thus, HIF-2α's function in ES cells was unclear. We performed DNA microarray analysis of hypoxia-inducible genes in WT, Hif-1α−/−, and Hif-2α−/− ES cells and identified 60 identical hypoxia-inducible genes in either WT or Hif-2α−/− ES cells. The majority of those tested were confirmed by Northern blot analysis. Besides several known hypoxia-inducible genes, a large number of novel hypoxia-inducible genes, such as Upp, Ak4, Glk, and Flnb, were also O2 regulated in the WT and Hif-2α−/− ES cells (Table 1). Induction in the HIF-1α-expressing WT and Hif-2α−/− ES cells but not in Hif-1α−/− ES cells suggests they are true HIF target genes.
Our DNA microarray study clearly shows that HIF-1α regulates a large number of target genes in ES cells. However, to our surprise, HIF-2α is not functional in ES cells. This conclusion is based on several lines of evidence. First, we determined that Hif-1α−/− ES cells express full-length HIF-2α mRNA and are devoid of HIF-2α isoforms (Fig. 5A). Next, we demonstrated that Hif-1α−/− ES cells exhibit no HIF-mediated hypoxic gene induction by investigating endogenous HIF-2α target genes as well as more-sensitive HRE reporters under a variety of stimuli, including severe hypoxia and several hypoxia mimetics. In addition, low levels of induction of several genes, including Vegf, Anaxa-2, and Ldha, observed in Hif-1α−/− ES cells is due to HIF-independent regulation, since similar induction is observed in Arnt−/− ES cells. Finally, HIF-2α lacks transcriptional activity in ES cells. If all the hypoxia-inducible genes observed in the WT or Hif-2α−/− ES cells were HIF-1α unique target genes, then we could not conclude that HIF-2α is inactive here. We previously showed that HIF-2α regulates genes such as Vegf, Glut-1, Adrp, Adm, and Ndrg-1 in 786-O cells, and these genes are also induced by HIF-1α in ES cells, as shown in Fig. 2 and Table 1, indicating they are HIF-1α/HIF-2α common target genes. Indeed, these genes can be induced by HIF-2α when HIF-2α becomes functional in ES cells (Fig. 8), while the HIF-1α unique target genes Pgk and Ldha remain unresponsive to overexpressed HIF-2α in ES cells (data not shown). Induction of the HIF-1α/HIF-2α common target genes Vegf, Glut-1, Adrp, Adm, and Ndrg-1 exclusively by HIF-1α but not by HIF-2α demonstrates that endogenous HIF-2α does not regulate its target genes in ES cells.
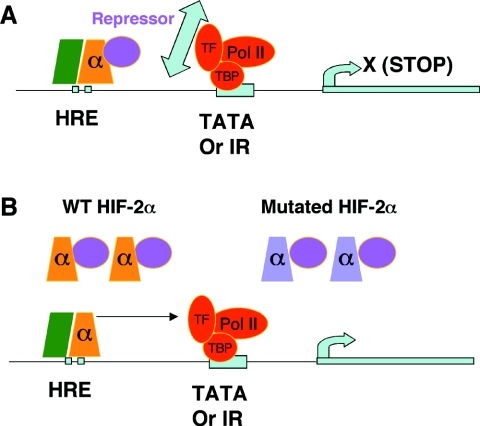
Having confirmed that the WT HIF-2α protein is expressed and binds to target gene promoters, we hypothesized that HIF-2α inhibition in ES cells occurs at the step of transcriptional cofactor recruitment. Although ES cells express levels of the HIF-2α protein similar to or even higher than that expressed by 786-O WT8 cells (as determined using an antibody generated from a peptide conserved between human and mouse HIF-2α) (Fig. 5B), this endogenous level of the HIF-2α protein is sufficient to induce target gene expression in 786-O cells but not in Hif-1α−/− ES cells. This difference suggests either that ES cells express a HIF-2α-specific transcriptional repressor or that they lack a HIF-2α-specific activator. Restoration of HIF-2α function in ES cells by overexpressed WT HIF-2α, particularly by the HIF-2α mutant, but not by the similar HIF-1α mutant suggests the presence of a HIF-2α-specific transcriptional repressor in these cells. Partial restoration of endogenous HIF-2α function by the HIF-2α mutant is likely due to its having both a positive effect in relieving repressor binding to endogenous HIF-2α and a negative effect by sequestering other transcriptional coactivators from endogenous HIF-2α. This experiment also proves that ES cells produce a functional HIF-2α transcript. Based on these data, we propose the following model: in ES cells, HIF-2α/ARNT dimers occupy target gene HREs but fail to activate transcription due to the expression of a HIF-2α-specific transcriptional corepressor. We hypothesize that this repressor inhibits the function of HIF-2α in ES cells by preventing HIF-2α from interacting with general transcriptional factors on the promoters. Overexpressed WT or a DNA-binding-defective HIF-2α protein serves as a sink for the repressor, allowing the repressor-free HIF-2α protein to stimulate target gene expression (Fig. 10). While our experiments collectively provided strong evidence for a repressor, definitive proof will ultimately come from the “rescue” of HIF-2α function in ES cells upon siRNA knock-down of the repressor.
FIG. 10.
Proposed model for HIF-2α inhibition in ES cells. (A) HIF-2α/HIF-1β(ARNT) dimers bind target gene HREs but fail to stimulate their transcription, likely due to inhibition of HIF-2α interaction with general transcription machinery by a HIF-2α-specific repressor. (B) Introduction of the WT or mutated HIF-2α protein renders HIF-2α active by titrating out the repressor. The green box represents the HIF-β (ARNT) subunit.
The HIF-1α and HIF-2α proteins exhibit a high degree of homology in their bHLH and PAS regions, domains important for DNA binding and ARNT dimerization. However, the two proteins exhibit limited similarities in their C termini, particularly the inhibitory domains and N-terminal transactivation domains. The structural diversity provides a molecular basis for their unique transcriptional cofactor requirements. For example, the NF-κB essential modulator (NEMO) has been shown to exclusively interact with HIF-2α and to promote its transcriptional activity by enhancing binding to CBP/p300 (3). NEMO is a coactivator for HIF-2α and is expressed in ES cells (according to DNA microarray data), indicating that NEMO is not the explanation for lack of HIF-2α function in ES cells. Another example of a HIF-2α-specific regulator is a proposed cytoplasmic protein in MEFs that prevents HIF-2α translocation to the nucleus (35). However, a recent report indicates that HIF-2α is not expressed in some MEF isolates (3). We also failed to detect HIF-2α protein in our primary MEF cultures (data not shown). Since HIF-2α in ES cells exhibits normal pVHL regulation and nuclear translocation, the putative HIF-2α repressor in ES cells is different from that in MEFs. CITED-2 and its isoform p35srj have been shown to inhibit transcriptional activity of HIF-1α by preventing coactivators p300/CBP and SRC-1 from binding to the HIF-1α C-terminal transactivation domain (2, 6, 23, 40). We determined that p300, SRC-1, and CITED-2 are expressed in ES cells (see Fig. S2 in the supplemental material). Expression of CITED-2 might explain HIF-2α inactivity in ES cells if CITED-2 has stronger inhibitory effects on HIF-2α. However, transfection of a CITED-2 expression plasmid did not prevent either HIF-1α and HIF-2α induction of cotransfected HRE-mediated reporter genes in ES cells, although CITED-2 inhibited HIF-1α and HIF-2α similarly in HEK293 cells (see Fig. S2 in the supplemental material). Thus, misexpression of coactivators p300 and SRC-1 or expression of CITED-2 is not involved in HIF-2α's inability to regulate its target genes in ES cells.
ChIP assays demonstrate that endogenous HIF-2α/ARNT dimers bind to the HREs of the HIF target genes Glut-1 and Vegf. Interestingly, the binding of HIF-1α (or HIF-2α) to the HREs is not enhanced by deletion of its counterpart α subunit in ES cells, suggesting that HIF-1α and HIF-2α might not bind the identical DNA elements. This is in accordance with the fact that HREs from Glut-1 and Vegf contain at least two potential binding sites, a conventional HIF-binding site (CGTG) and an inverted repeat (CACG for Glut-1 and CAGG for Vegf) (24). It is worth noting that the HIF-1α protein induces Glut-1 and Vegf expression to similar levels in both Hif-2α−/− and WT ES cells, consistent with the idea that HIF-2α DNA binding in WT ES cells does not interfere with HIF-1α function. A more direct assay, such as in vivo DNA footprinting, is required to determine whether the above-described phenomenon is the consequence of unsaturated binding or occupying unique HIF binding sites on the promoters.
The discovery that some HIF-2α-expressing cells may not harbor a functional HIF-2α protein helps to resolve some confusion about the role of HIF-2α in hypoxic responses. It is now clear that loss of HIF-1α abolishes all HIF-dependent hypoxic gene induction in ES cells, since HIF-2α is not functional in these cells. Furthermore, our data also indicate that expression of HIF-2α mRNA and protein does not guarantee HIF-2α activity; thus, a meaningful study of target gene specificity using siRNA technology to specifically knock down HIF-1α or HIF-2α must use cells where both HIF-1α and HIF-2α are active.
The tumor-promoting effects of VHL mutations appear to be dependent on HIF-2α-mediated target gene regulation. Thus, lack of HIF-2α activity might block this effect of VHL mutations. HIF-2α has been shown to play a critical role in tumor formation by pVHL-deficient RCC cells, and HIF-2α is clearly functional in these cells (16, 50, 53). Nonfunctional HIF-2α in ES cells might be partially responsible for our observation that pVHL-deficient ES cells fail to promote tumor growth in nude mice, despite the fact that these ES cells display constitutive HIF-1α and HIF-2α protein and HIF-1α target gene expression, as observed for pVHL-deficient RCC cells (29, 37). In agreement with the teratoma model, fibrosarcomas derived from Vhl−/− MEFs are significantly smaller than WT controls (28), and MEFs appear to harbor no HIF-2α activity due to cytoplasmic trapping (35) or lack of expression (3). In contrast to Vhl−/− ES cells, ES cell-derived teratomas with targeted replacement of HIF-1α by HIF-2α display a proliferative advantage in comparison to WT ES cells (8). Increased teratoma expression of HIF-2α target genes, such as Vegf, transforming growth factor α (TGF-α), and cyclin D1, suggests that HIF-2α is functional in these cells, presumably by overcoming a HIF-2α repressor through increased expression of the HIF-2α protein. Although VHL and HIF-2α are broadly expressed, human VHL mutations give rise to only a few tissue-specific tumors. Expression of a HIF-2α-specific repressor might play a role in limiting the tissue penetration of VHL mutations. It will be interesting to identify the HIF-2α repressor and to see whether there is a negative correlation between tissues that express the HIF-2α repressor and risk levels of VHL disease.
Recently, we and others showed that HIF-2α regulates TGF-α and the Pou transcription factor Oct-4 (also known as Oct-3/4 and Pou5F1) in RCC cells (16, 38, 53). Moreover, these genes are specifically regulated by HIF-2α in RCC cells (38). Since HIF-2α is expressed and its protein is stabilized in hypoxic ES cells, the inhibition of HIF-2α target gene expression by a HIF-2α transcriptional repressor may be required for proper ES cell function. Expression of Oct-4 in undifferentiated ES cells is essential for maintaining stem cell pluripotency (33). When Oct-4 expression is reduced or increased, ES cells lose pluripotency and start differentiating, demonstrating the importance of maintaining correct Oct-4 expression levels (34). We recently observed that expanded HIF-2α expression in ES cells by HIF-2α knock-in at the Hif-1α locus leads to severe developmental defects in mouse embryos, defective hematopoetic stem cell differentiation in embryoid bodies, and larger teratomas (K. L. Covello et al., submitted for publication). Interestingly, we found that enhanced Oct-4 expression in these cells is partially responsible for these defects, providing strong evidence for the tight regulation of HIF-2α activity in ES cells and early embryonic development.
Our results demonstrated that HIF-2α is not functional in some cell types, such as ES cells. However, HIF-1α appears to be functional in all cells tested. This might reflect their distinct expression patterns and functions. For example, HIF-1α is present in organisms from Caenorhabditis elegans to humans, while HIF-2α exists only in more complicated vertebrate species, such as chicken, quail, and mammals (9, 10, 20). In addition, HIF-1α is universally expressed while HIF-2α expression is more tissue restricted, even in organisms where both isoforms are expressed. From a functional standpoint, HIF-1α appears to regulate genes involved in basic cellular activity, like the glycolytic pathway, while HIF-2α is more involved in genes having special function such as Oct-4.
In summary, we demonstrate that HIF-1α, but not HIF-2α, is functional in ES cells, showing a clear regulatory difference between HIF-1α and HIF-2α. While hypoxic stabilization of HIF-α is a critical regulatory step for HIF transcriptional activity, stabilization of HIF-2α is insufficient for transcriptional activity. Furthermore, we provide compelling evidence for a model whereby HIF-2α's inability to regulate its target genes in ES cells is due to the expression of a HIF-2α-specific corepressor that globally inactivates its function. Identification of such a novel regulatory mechanism is critical to understanding the role of HIF-2α in hypoxic responses and in tumorigenesis and provides a possible explanation for VHL disease tissue specificity.
Supplementary Material
Acknowledgments
We thank members of the Simon lab for thoughtful discussions and reading of the manuscript.
This work was supported by grant 66130 (M.C.S.) from the National Institutes of Health, the Howard Hughes Medical Institute, and the Abramson Family Cancer Research Institute. M.C.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adelman, D. M., M. Gertsenstein, A. Nagy, M. C. Simon, and E. Maltepe. 2000. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 14:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya, S., C. L. Michels, M. K. Leung, Z. P. Arany, A. L. Kung, and D. M. Livingston. 1999. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 13:64-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bracken, C. P., M. L. Whitelaw, and D. J. Peet. 2005. Activity of hypoxia-inducible factor 2 alpha is regulated by association with the NF-kappa B essential modulator. J. Biol. Chem. 280:14240-14251. [DOI] [PubMed] [Google Scholar]
- 4.Brusselmans, K., F. Bono, P. Maxwell, Y. Dor, M. Dewerchin, D. Collen, J. M. Herbert, and P. Carmeliet. 2001. Hypoxia-inducible factor-2alpha (HIF-2alpha) is involved in the apoptotic response to hypoglycemia but not to hypoxia. J. Biol. Chem. 276:39192-39196. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, E. Keshert, and E. Keshet. 1998. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 6.Carrero, P., K. Okamoto, P. Coumailleau, S. O'Brien, H. Tanaka, and L. Poellinger. 2000. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol. Cell. Biol. 20:402-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Compernolle, V., K. Brusselmans, T. Acker, P. Hoet, M. Tjwa, H. Beck, S. Plaisance, Y. Dor, E. Keshet, F. Lupu, B. Nemery, M. Dewerchin, P. Van Veldhoven, K. Plate, L. Moons, D. Collen, and P. Carmeliet. 2002. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat. Med. 8:702-710. [DOI] [PubMed] [Google Scholar]
- 8.Covello, K. L., M. C. Simon, and B. Keith. 2005. Targeted replacement of hypoxia-inducible factor-1alpha by a hypoxia-inducible factor-2alpha knock-in allele promotes tumor growth. Cancer Res. 65:2277-2286. [DOI] [PubMed] [Google Scholar]
- 9.Elvert, G., S. Lanz, A. Kappel, and I. Flamme. 1999. mRNA cloning and expression studies of the quail homologue of HIF-2alpha. Mech. Dev. 87:193-197. [DOI] [PubMed] [Google Scholar]
- 10.Favier, J., H. Kempf, P. Corvol, and J. M. Gasc. 1999. Cloning and expression pattern of EPAS1 in the chicken embryo. Colocalization with tyrosine hydroxylase. FEBS Lett. 462:19-24. [DOI] [PubMed] [Google Scholar]
- 11.Giaccia, A. J., M. C. Simon, and R. Johnson. 2004. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 18:2183-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giffard, R. G., L. Xu, H. Zhao, W. Carrico, Y. Ouyang, Y. Qiao, R. Sapolsky, G. Steinberg, B. Hu, and M. A. Yenari. 2004. Chaperones, protein aggregation, and brain protection from hypoxic/ischemic injury. J. Exp. Biol. 207:3213-3220. [DOI] [PubMed] [Google Scholar]
- 13.Gothie, E., D. E. Richard, E. Berra, G. Pages, and J. Pouyssegur. 2000. Identification of alternative spliced variants of human hypoxia-inducible factor-1alpha. J. Biol. Chem. 275:6922-6927. [DOI] [PubMed] [Google Scholar]
- 14.Gubits, R. M., R. E. Burke, G. Casey-McIntosh, A. Bandele, and F. Munell. 1993. Immediate early gene induction after neonatal hypoxia-ischemia. Brain Res. Mol. Brain Res. 18:228-238. [DOI] [PubMed] [Google Scholar]
- 15.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 16.Hu, C. J., L. Y. Wang, L. A. Chodosh, B. Keith, and M. C. Simon. 2003. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 23:9361-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iliopoulos, O., A. Kibel, S. Gray, and W. G. Kaelin, Jr. 1995. Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1:822-826. [DOI] [PubMed] [Google Scholar]
- 18.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain, S., E. Maltepe, M. M. Lu, C. Simon, and C. A. Bradfield. 1998. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech. Dev. 73:117-123. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, H., R. Guo, and J. A. Powell-Coffman. 2001. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl. Acad. Sci. USA 98:7916-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, K. D., and E. H. Bresnick. 2002. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 26:27-36. [DOI] [PubMed] [Google Scholar]
- 22.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 23.Kallio, P. J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura, H., A. Weisz, T. Ogura, Y. Hitomi, Y. Kurashima, K. Hashimoto, F. D'Acquisto, M. Makuuchi, and H. Esumi. 2001. Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J. Biol. Chem. 276:2292-2298. [DOI] [PubMed] [Google Scholar]
- 25.Kondo, K., W. Y. Kim, M. Lechpammer, and W. G. Kaelin, Jr. 2003. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLOS Biol. 1:E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo, K., J. Klco, E. Nakamura, M. Lechpammer, and W. G. Kaelin, Jr. 2002. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1:237-246. [DOI] [PubMed] [Google Scholar]
- 27.Lando, D., I. Pongratz, L. Poellinger, and M. L. Whitelaw. 2000. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor. J. Biol. Chem. 275:4618-4627. [DOI] [PubMed] [Google Scholar]
- 28.Mack, F. A., J. H. Patel, M. P. Biju, V. H. Haase, and M. C. Simon. 2005. Decreased growth of Vhl−/− fibrosarcomas is associated with elevated levels of cyclin kinase inhibitors p21 and p27. Mol. Cell. Biol. 25:4565-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mack, F. A., W. K. Rathmell, A. M. Arsham, J. Gnarra, B. Keith, and M. C. Simon. 2003. Loss of pVHL is sufficient to cause HIF dysregulation in primary cells but does not promote tumor growth. Cancer Cell 3:75-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maltepe, E., J. V. Schmidt, D. Baunoch, C. A. Bradfield, and M. C. Simon. 1997. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature 386:403-407. [DOI] [PubMed] [Google Scholar]
- 31.Maranchie, J. K., J. R. Vasselli, J. Riss, J. S. Bonifacino, W. M. Linehan, and R. D. Klausner. 2002. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 1:247-255. [DOI] [PubMed] [Google Scholar]
- 32.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 33.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 34.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 35.Park, S. K., A. M. Dadak, V. H. Haase, L. Fontana, A. J. Giaccia, and R. S. Johnson. 2003. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1α (HIF-1α): role of cytoplasmic trapping of HIF-2α. Mol. Cell. Biol. 23:4959-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng, J., L. Zhang, L. Drysdale, and G. H. Fong. 2000. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. USA 97:8386-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathmell, W. K., M. M. Hickey, N. A. Bezman, C. A. Chmielecki, N. C. Carraway, and M. C. Simon. 2004. In vitro and in vivo models analyzing von Hippel-Lindau disease-specific mutations. Cancer Res. 64:8595-8603. [DOI] [PubMed] [Google Scholar]
- 38.Raval, R. R., K. W. Lau, M. G. Tran, H. M. Sowter, S. J. Mandriota, J. L. Li, C. W. Pugh, P. H. Maxwell, A. L. Harris, and P. J. Ratcliffe. 2005. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25:5675-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sang, N., J. Fang, V. Srinivas, I. Leshchinsky, and J. Caro. 2002. Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1 alpha is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol. Cell. Biol. 22:2984-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schofield, C. J., and P. J. Ratcliffe. 2004. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell. Biol. 5:343-354. [DOI] [PubMed] [Google Scholar]
- 42.Scortegagna, M., K. Ding, Y. Oktay, A. Gaur, F. Thurmond, L. J. Yan, B. T. Marck, A. M. Matsumoto, J. M. Shelton, J. A. Richardson, M. J. Bennett, and J. A. Garcia. 2003. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-/- mice. Nat. Genet. 35:331-340. [DOI] [PubMed] [Google Scholar]
- 43.Scortegagna, M., K. Ding, Q. Zhang, Y. Oktay, M. J. Bennett, M. Bennett, J. M. Shelton, J. A. Richardson, O. Moe, and J. A. Garcia. 2005. HIF-2alpha regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood 105:3133-3140. [DOI] [PubMed] [Google Scholar]
- 44.Semenza, G. L. 2000. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 88:1474-1480. [DOI] [PubMed] [Google Scholar]
- 45.Shima, D. T., U. Deutsch, and P. A. D'Amore. 1995. Hypoxic induction of vascular endothelial growth factor (VEGF) in human epithelial cells is mediated by increases in mRNA stability. FEBS Lett. 370:203-208. [DOI] [PubMed] [Google Scholar]
- 46.Tian, H., R. E. Hammer, A. M. Matsumoto, D. W. Russell, and S. L. McKnight. 1998. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 12:3320-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 48.Wang, G. L., and G. L. Semenza. 1995. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270:1230-1237. [DOI] [PubMed] [Google Scholar]
- 49.Wang, V., D. A. Davis, M. Haque, L. E. Huang, and R. Yarchoan. 2005. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 65:3299-3306. [DOI] [PubMed] [Google Scholar]
- 50.Warnecke, C., Z. Zaborowska, J. Kurreck, V. A. Erdmann, U. Frei, M. Wiesener, and K. U. Eckardt. 2004. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J. 18:1462-1464. [DOI] [PubMed] [Google Scholar]
- 51.Wenger, R. H., A. Rolfs, P. Spielmann, D. R. Zimmermann, and M. Gassmann. 1998. Mouse hypoxia-inducible factor-1alpha is encoded by two different mRNA isoforms: expression from a tissue-specific and a housekeeping-type promoter. Blood 91:3471-3480. [PubMed] [Google Scholar]
- 52.Wiesener, M. S., J. S. Jurgensen, C. Rosenberger, C. K. Scholze, J. H. Horstrup, C. Warnecke, S. Mandriota, I. Bechmann, U. A. Frei, C. W. Pugh, P. J. Ratcliffe, S. Bachmann, P. H. Maxwell, and K. U. Eckardt. 2003. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J. 17:271-273. [DOI] [PubMed] [Google Scholar]
- 53.Wykoff, C. C., C. Sotiriou, M. E. Cockman, P. J. Ratcliffe, P. Maxwell, E. Liu, and A. L. Harris. 2004. Gene array of VHL mutation and hypoxia shows novel hypoxia-induced genes and that cyclin D1 is a VHL target gene. Br J. Cancer 90:1235-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.