Figure 3.—
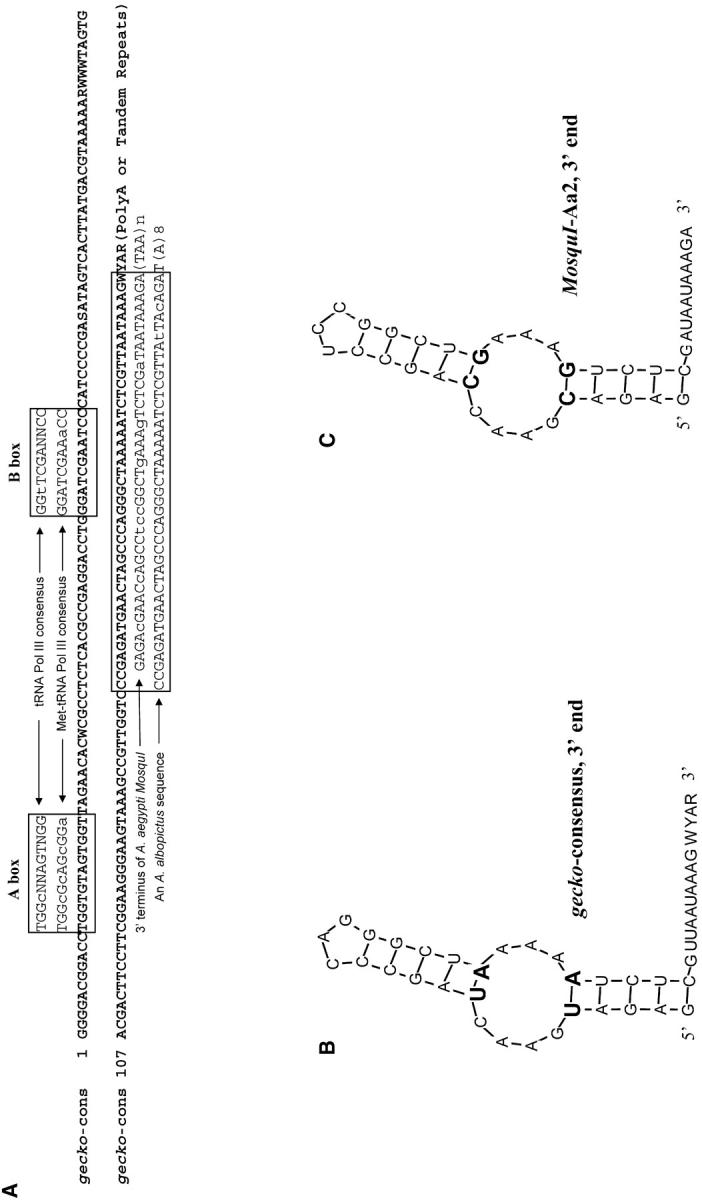
(A) Consensus of A. aegypti gecko and its features. 5′ gecko sequences were aligned to the A and B boxes of polymerase III promoters that are derived from the consensus sequences of tRNA Pol III and Met-tRNA Pol III (Deininger 1989). Thirty-three base pairs of the 41-bp fragment at the 3′ end of gecko are identical to the 3′ terminus of MosquI, a non-LTR retrotransposon in A. aegypti (Tu and Hill 1999). Forty-two base pairs of the 44-bp fragment at the 3′ end of gecko are identical to an uncharacterized sequence in A. albopictus (GenBank AF144549). Uppercase letters indicate conservation between gecko and the aligned sequences. Lowercase letters indicate variations. Twenty base pairs of a 21-bp region near the 5′ end of gecko (nucleotides 25–45 in the consensus) is identical to the reverse strand of a yeast tRNA sequence (Suzuki et al. 1994), which is not shown. (B) Predicted secondary structure of the 3′ end of the gecko consensus as shown in Figure 1D. (C) Predicted secondary structure of the 3′ end of MosquI-Aa2, a full-length copy of a non-LTR retrotransposon in A. aegypti. (B and C) Structures predicted using the GeneQuest program of Lasergene. Mfold of GCG was also used, which gave similar structural predictions. The two pairs of complementary changes between structures in B and C are in boldface type and a larger type size.
