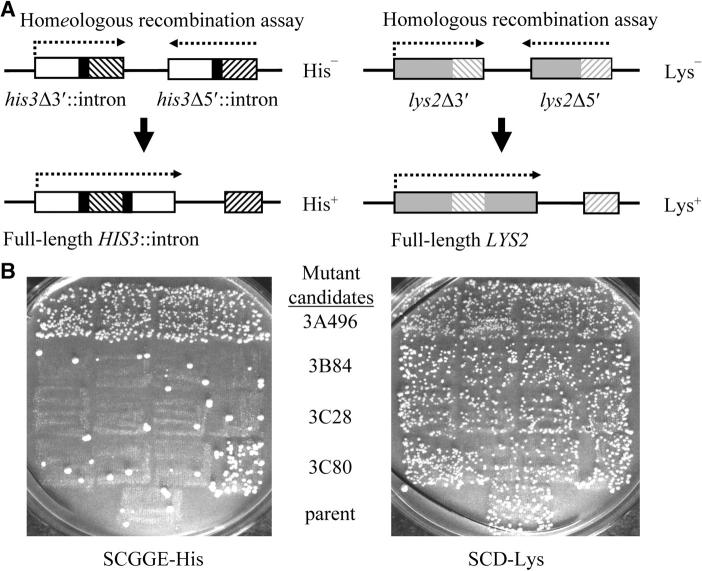
Figure 1.—
Inverted repeat assay. (A) Homeologous recombination was assayed using 91% identical cβ2/cβ7 inverted repeats (hatching) fused to intron splice sites (solid boxes) and placed next to the 5′ and 3′ halves of the HIS3 gene (open boxes). Recombination between the repeats that leads to reorientation of the intervening sequence reconstitutes a full-length HIS3 gene and results in a His+ phenotype. Homologous recombination was assayed using overlapping 5′ and 3′ portions of the LYS2 gene (shaded boxes), thus generating 100%-identical repeats in an inverted orientation (shaded hatching). Recombination between the repeats that leads to reorientation of the intervening sequence reconstitutes a full-length LYS2 gene and results in a Lys+ phenotype. Quantitative measurements of homologous recombination could also be measured in strains containing 100%-identical cβ2/cβ2 repeats fused to HIS3 sequences. Similar effects on homologous recombination are seen with the 100% identical LYS2 substrates and the 100% identical HIS3 substrates. (B) Screen for mutants defective in the regulation of homeologous recombination. Representative plates from retests of mutant candidates are shown. The unmutagenized parent (bottom square) and four purified isolates of each mutant candidate were patched (row of four squares) onto YEPD and then replica plated to SDGGE-His (left) and SCD-Lys (right). Prototrophic papillae represent recombinants. Whereas homeologous recombination (His+) is suppressed relative to homologous recombination (Lys+) in the parent strain, only candidate 3A496 exhibited a consistent elevation in homeologous recombination relative to homologous recombination and was later determined to contain a mutation in MSH6.