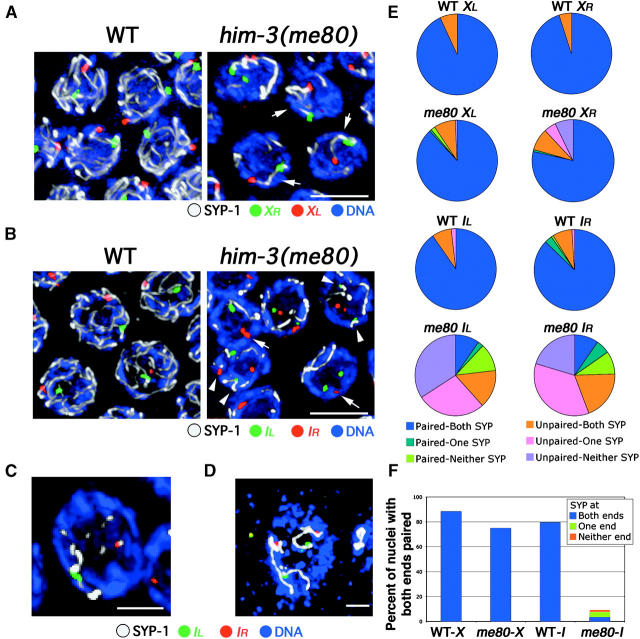
Figure 3.—
Simultaneous assessment of pairing and synapsis for chromosomes X and I. Analysis of FISH for two loci (red and green) in conjunction with SYP-1 immunostaining (white) on DAPI-stained chromosomes (blue) in mid/late-pachytene nuclei from WT and him-3(me80) hermaphrodites is shown. Images in A–C show nuclei from whole-mount germ lines, and the image in D shows a nucleus from a squash preparation; all images are projections of 3-D data stacks encompassing whole nuclei. (A) FISH for two loci at opposite ends of the X chromosome. Both XL and XR are usually paired and associated with a single continuous stretch of SYP-1 immunostaining (arrows) in the him-3(me80) mutant. Bar, 5 μm. (B–D) FISH for two loci at opposite ends of chromosome I. In B, arrows indicate examples of nuclei in the him-3(me80) mutant where IR FISH signals are paired but are not associated with a stretch of SYP-1; arrowheads indicate examples where unpaired FISH signals associate with SYP-1 stretches. (C) A him-3(me80) nucleus in which the FISH signals at both ends of chromosome I are paired, but are associated with different SYP-1 stretches. (D) A him-3(me80) nucleus in which all four chromosome I FISH signals are unpaired yet are associated with SYP-1 stretches. Bars: B and D, 5 μm; C, 2 μm. (E) Pie charts showing simultaneous quantitation of pairing of FISH signals at a given locus and association of those FISH signals with a stretch of anti-SYP-1 immunostaining. Each pie chart represents data for a single probe in either wild type or the him-3(me80) mutant; between 102 and 143 nuclei were scored in each case. Nuclei were classified with respect to whether FISH signals for the given locus were paired (i.e., the distance between their peak signal intensities was ≤0.7 μm), and nuclei were further classified according to whether both, only one, or neither of the FISH signals for that locus were associated with (i.e., touched or overlapped with) a SYP-1 stretch. One classification requires further explanation: for all probes and all genotypes, a subset of nuclei was classified as “Unpaired-Both SYP.” For all four probes in WT, and for the X probes in him-3(me80), in the vast majority of these cases the distances between the FISH signals fell in the 0.8–1.0 μm range and both signals were associated with the same SYP-1 stretch. For the chromosome I probes in him-3(me80), in contrast, most nuclei in the Unpaired-Both SYP class have the FISH signals far apart (2.3 ± 1.0 μm, n = 50) and associated with distinct SYP-1 stretches. (F) Bar graph incorporating the data for the two FISH probes targeting opposite ends of the same chromosome. Graphs indicate the percentage of nuclei showing homologous pairing at both ends of the indicated chromosome; such nuclei are further classified with respect to whether both, only one, or neither of the two sets of paired FISH signals were associated with SYP-1 staining.