Figure 1.—
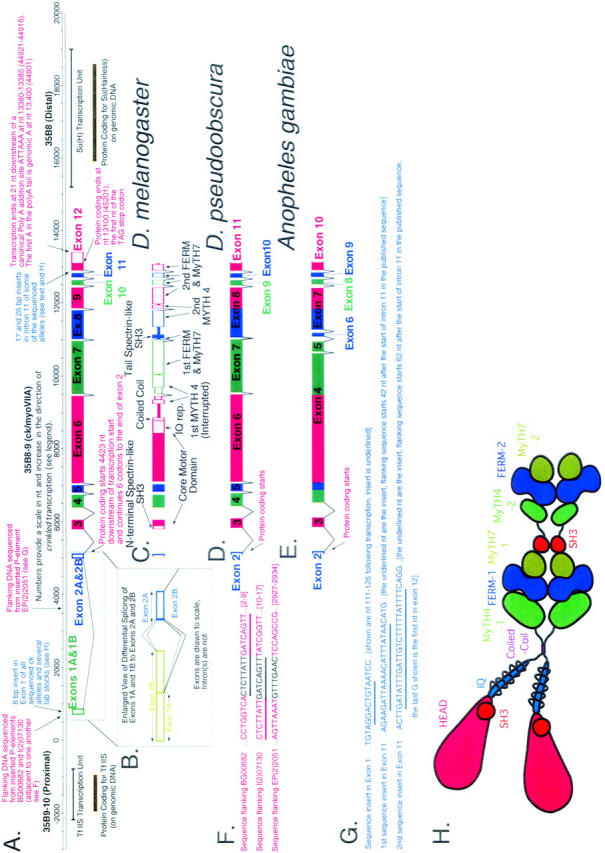
A schematic overview of genomic organization at the crinkled locus at polytene location 35B shows ck/myoVIIA transcription units from Drosophila melanogaster, D. pseudoobscura, and Anopheles gambiae and indicates significant differences in exon/intron structure. (A) Numbers provide a scale in nt and increase in the direction of crinkled transcription (“0” was chosen early in the project at a site predicted to be close to transcription start—its location is therefore arbitrary). Transcription start, identified by 5′ RACE, is at nt 663. This origin corresponds to nt 58301 in accession no. AE003646 from the Drosophila genome project (in which numbers decrease in the direction of ck transcription). Adjacent genes TfIIS and Suppressor of Hairless are also shown. (B) An enlarged view of exons 1 and 2 from D. melanogaster shows differential splicing at the first intron. (C) Domain structure of ck/myoVIIA protein mapped onto the exon/intron structure of the D. melanogaster gene. (D and E) Exons and introns in the D. pseudoobscura and A. gambiae ck/myoVIIA genes are shown (compare to A). These species last shared a common ancestor with D. melanogaster 25 million and 250 million years ago, respectively (Russo et al. 1995; Zdobnov et al. 2002). In D. pseudoobscura, a single exon 8 replaces exons 8 and 9 of D. melanogaster. In contrast, in A. gambiae, the exons corresponding to the melanogaster exons 4, 5, 6, and part of 7 are “fused” into exon 4. Likewise, parts of the melanogaster exons 8 and 9 are fused to make the A. gambiae exon 7. (F) Sequence flanking three P-element-induced alleles of ck. Sequence in black is the 8-bp target sequence that is duplicated upon P insertion. Note that the target sites for BG00682 and P(PZ)07130 are directly adjacent to one another (shared sequence is underlined). (G) Insertional polymorphisms in D. melanogaster exon 1 and intron 11. (H) Schematic of the ck/myoVIIA protein outlines the overall structure of the protein and is color coded using the scheme in Figure 2. The schematic is drawn approximately to scale: the area of each “domain” is roughly proportional to the number of amino acids in that domain. Contact between dimerized heavy chains is shown at the coiled-coil region, the FERM domains, and the tail SH3 domain because those domains are thought to mediate protein-protein interactions. It is important to note that no evidence, either for or against such interactions, exists. Similarly, the MyTH4 and MyTH7 domains may also contribute to intradimer interactions. Rings drawn around the IQ motif region represent light chains.
