Abstract
Fission yeast mutants defective in DNA replication have widely varying morphological phenotypes. We designed a screen for temperature-sensitive mutants defective in the process of replication regardless of morphology by isolating strains unable to rereplicate their DNA in the absence of cyclin B (Cdc13). Of the 42 rereplication-defective mutants analyzed, we were able to clone complementing plasmids for 10. This screen identified new alleles of the APC subunit cut9+, the initiation/checkpoint factor rad4+/cut5+, and the first mutant allele of psf2+, a subunit of the novel GINS replication complex. Other genes identified are likely to play general roles in gene expression and protein localization.
THE fission yeast Schizosaccaromyces pombe is an excellent system for analysis of DNA replication. With facile genetics and large origins of replication similar to those of larger eukaryotes, fission yeast has emerged over the last 10 years as a major system for understanding this fundamental biological event. These insights have relied on the analysis of an extensive collection of mutants defective in replication. Many of the S-phase mutants were isolated in the original cell division cycle (cdc) screen (Nasmyth and Nurse 1981); these mutants grow without dividing and arrest within one cell cycle. Others were identified for their cell untimely torn (cut) phenotype. These are generally checkpoint-defective initiation mutants, which bypass DNA replication and proceed directly into M phase (e.g., Saka and Yanagida 1993). However, similar cdc or cut phenotypes have been observed for a diverse group of mutants not involved in DNA replication, making screens based solely on morphology insufficient to isolate S-phase genes. Moreover, previous screens that isolated S-phase mutants in yeast were not saturating, because new replication genes continue to be identified through biochemical and molecular methods (e.g., Kanemaki et al. 2003; Takayama et al. 2003). Importantly, mutants affecting many of these new S-phase mutants do not result in a clear cdc or cut phenotype.
To identify mutants specifically defective in S-phase functions, we designed a screen based on the process of replication rather than on the terminal morphology of the mutant strain. We assessed the ability of mutant cells to undergo repeated rounds of S phase without an intervening mitosis, a phenomenon called rereplication. Genome-wide rereplication occurs when the activity of the G2/M phase form of the cyclin-dependent kinase Cdc2p is manipulated by mutation of cdc2 (Broek et al. 1991), overexpression of the Rum1p inhibitor (Moreno and Nurse 1994), or depletion of the B-type cyclin Cdc13p (Hayles et al. 1994; Fisher and Nurse 1996). Loss of S-phase genes abolishes the ability of the cells to rereplicate, suggesting that it relies on normal S-phase functions (Fisher and Nurse 1996; Snaith and Forsburg 1999). Once the rereplication mechanism is triggered, cells lose viability as they increase ploidy (Moreno and Nurse 1994); fission yeast does not tolerate levels of DNA much beyond diploidy (Molnar and Sipiczki 1993).
We identified new alleles of two known genes: the anaphase-promoting complex (APC) subunit cut9+ and the initiation/checkpoint protein rad4/cut5+. We also isolated the first mutant allele of the psf2+ gene, which encodes a likely subunit of the GINS (Go, Ichi, Nii, and San, or five, one, two, and three, respectively, in Japanese) replication complex, recently identified in Saccharomyces cerevisiae and Xenopus (Kanemaki et al. 2003; Kubota et al. 2003; Takayama et al. 2003). We identified a mutation in one new gene, dre4+ (dre, defects in rereplication), which has defects in S-phase progression, chromatin structure, and cytokinesis. Clones rescuing other dre mutants do not contain genes with obvious replication function, but instead encode likely candidates for RNA metabolism or protein trafficking.
MATERIALS AND METHODS
Strains and manipulations:
Strains used in this study are listed in Table 1. Strain FY875 (Snaith and Forsburg 1999) and its derivatives were maintained on thiamine-free Edinburgh minimal media (EMM) with appropriate supplements, and strains with no rereplication background were maintained on YES (yeast extract plus supplements) agar plates using standard techniques (Moreno et al. 1991). Matings were performed on synthetic sporulation agar (SPA; Gutz et al. 1974) plates for 2–3 days at 25°. Transformations were carried out by electroporation (Kelly et al. 1993). For nitrogen starvation, cells were grown to midlog phase in thiamine-free EMM, washed twice in nitrogen-free EMM, inoculated into fresh nitrogen-free EMM plus 7.5 μg/ml adenine, and starved for 16 hr at 25°. For asynchronous temperature-shift analyses, cells were grown to OD595 = 0.4 and incubated at 25° and 36° for the indicated times. Strain FY255 (Table 1) was used to backcross the rereplication mutant candidates and isolate the temperature-sensitive (ts) mutation.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FY254 | h− ura4-D18 leu1-32 ade6 | Our stock |
| FY255 | h+ ura4-D18 leu1-32 ade6 | Our stock |
| FY261 | h+ura4-D18 leu1-32 ade6 | Our stock |
| FY421 | h− Δchk1::ura4+ura4-D18 leu1-32 ade6 | T. Carr |
| FY865 | h− Δcds1::ura4+ ura4-D18 leu1-32 | D. Griffiths |
| FY875 | h− Δcdc13::ura4+leu1-32::p[nmt*. cdc13+-leu1+]ura4-D18 ade6 | Our stock |
| FY1068 | h+cut9-665 ura4-D18 leu1-32 ade6 | Our stock |
| FY1107 | h− Δrad3::ura4+ ura4-D18 leu1-32 ade6 | Our stock |
| FY1114 | h−rad4-116 ura4-D18 leu1-32 ade6 | Our stock |
| FY1304 | h− cut9-41 ade6 | This study |
| FY1305 | h+ cut9-665 ade6 | Our stock |
| FY2711 | h+ psf2-209 ura4-D18 leu1-32 ade6 | This study |
| FY2712 | h− psf2-209 ura4-D18 leu1-32 ade6 | This study |
| FY2958 | h+ rad4-42 ura4-D18 leu1-32 ade6 | This study |
| FY2959 | h+ dre6-82 ura4-D18 leu1-32 ade6 | This study |
| FY2961 | h+ dre10-54 ura4-D18 leu1-32 ade6 | This study |
| FY2963 | h+ dre11-56 ura4-D18 leu1-32 ade6 | This study |
| FY2964 | h+ dre12-195 ura4-D18 leu1-32 ade6 | This study |
| FY2966 | h+ dre14-234 ura4-D18 leu1-32 ade6 | This study |
| FY2967 | h+ dre15-21 ura4-D18 leu1-32 ade6 | This study |
| FY2969 | h+ dre16-38 ura4-D18 leu1-32 ade6 | This study |
| FY2971 | h+ dre19-6 ura4-D18 leu1-32 ade6 | This study |
| FY2972 | h+ dre20-16 ura4-D18 leu1-32 ade6 | This study |
| FY2973 | h+ dre21-3 ura4-D18 leu1-32 ade6 | This study |
| FY2975 | h+ dre22-21 ura4-D18 leu1-32 ade6 | This study |
| FY2977 | h− dre23-34 ura4-D18 leu1-32 ade6 | This study |
| FY2978 | h+ dre24-8 ura4-D18 leu1-32 ade6 | This study |
| FY2979 | h+ dre25-16 ura4-D18 leu1-32 ade6 | This study |
| FY3033 | h−/h− dre24-8 (UV6-8) Δcdc13::ura4+ leu1-32[nmt*-cdc13+ leu1+] ura4-D18 ade6 | This study |
Isolation of ts mutants:
For ultraviolet (UV) mutagenesis, FY875 was grown to OD595 = 0.8 in thiamine-free EMM plus supplements, 1000 cells were plated and exposed to 200 J/m2 UV light in a Stratalinker 2400 (Stratagene, La Jolla, CA), resulting in a 50% killing. The protocol described in Moreno et al. (1991) was followed for 1-methyl-3 nitro-1 nitrosoguanidine (70-25-7, Sigma-Aldrich, St. Louis) mutagenesis of FY875. Mutagenized cells were incubated at 25° for 5 days, replica plated to phloxin B, and incubated at 36° for 2 days. Temperature-sensitive colonies were identified, streaked out at least twice, and frozen down. All temperature-sensitive strains were tested for their ability to rereplicate on plus-thiamine medium at 36° to make sure that the ts mutation was not in the nmt promoter.
Identification of rereplication mutants in screening protocol:
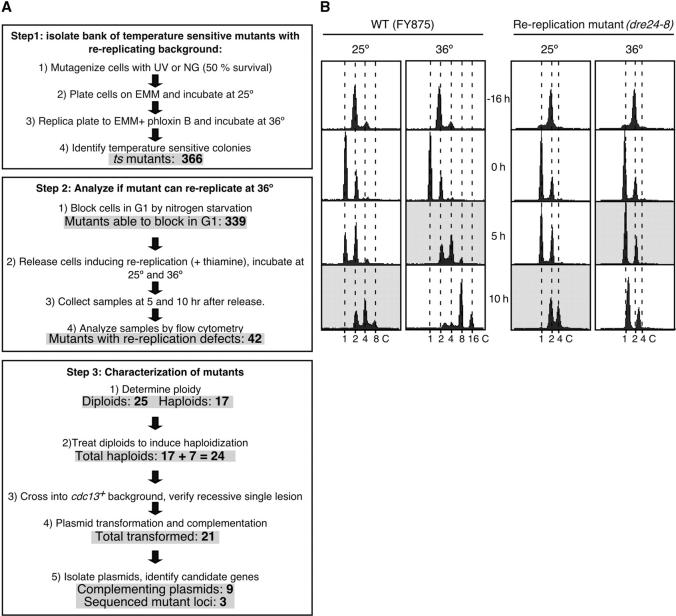
The isolated temperature-sensitive strains were arrested in G1 by nitrogen starvation. The cultures were divided in two and released into the cell cycle by adding nitrogen and thiamine to turn the nmt promoter off and induce rereplication. One culture was placed at 25° and the other at 36°. Samples were collected after 5 and 10 hr, ethanol fixed, and analyzed by flow cytometry as described previously (Gomez et al. 2002). Flow cytometry profiles corresponding to 10 hr at 25° (rereplication positive control) and 5 hr at 36° were compared (Figure 2B, shaded background profiles), and strains with different profiles were identified as rereplication mutant candidates.
Figure 2.—
(A) Scheme showing the steps and results of the screening method approach. (B) Example of interesting rereplication mutant isolated with the screening approach. Strains FY875 and FY3033 (rereplication candidate) were arrested in G1 by nitrogen starvation. Cultures were divided in two and released by adding nitrogen and thiamine. One culture was placed at 25° and the other at 36°. Samples were collected after 5 and 10 hr, ethanol fixed, and analyzed by flow cytometry. Flow cytometry profiles corresponding to 10 hr at 25° (rereplication positive control) and to 5 hr at 36° were compared (shaded background profiles). DNA content is indicated at the bottom.
Complementation of mutations by plasmid clones:
Cells were grown at 25° and transformed with a fission yeast genomic DNA library (generous gift of T. Carr). Transformants were plated on EMM lacking uracil to select for the plasmid, incubated for 24 hr at 25°, and shifted to 36° for 3 days. Plasmid-suppressed colonies were streaked out twice on EMM lacking uracil and the library vectors were recovered. All plasmids were retransformed into the corresponding ts strain to confirm the suppression and sequenced. Primers to amplify the complete open reading frame of some candidate genes were designed and used in PCR amplifications. Genomic DNA of the corresponding mutants and a wild-type strain was used as template. All PCRs were performed in duplicate, and DNA products were cloned and sequenced. Sequence of oligonucleotides use in PCRs and sequencing reactions are available upon request.
Haploidization of h−/h− diploids:
The m-fluorophenylalanine (m-FPA; F-5162, Sigma-Aldrich, St. Louis) haploidization protocol described in Kohli et al. (1977) was used with some modifications. Cells were streaked out on EMM + 0.1% m-FPA and incubated at 25° for 5 days. Cells were then suspended in H2O, and 500 were plated on EMM + supplements + phloxin B and incubated at 25°. Haploid colonies were distinguished from diploid colonies by their pale pink color and smaller cell size. Putative haploids were streaked out and analyzed by flow cytometry. The m-FPA method was inefficient so we employed a tetraploidization approach. A wild type h+/h+ diploid was isolated by spontaneous diploidization of strain FY261 and mated to our h−/h− rereplication temperature-sensitive mutants. Spores were plated on YES and incubated for 4 days at 25°. Colonies were replica plated to YES + phloxin B and incubated at 36° and to SPA to analyze their sporulation competence. Temperature-sensitive diploid colonies that formed spores on SPA plates were isolated and induced to sporulate. Spores were plated, and temperature-sensitive colonies were isolated and analyzed by flow cytometry.
DNA staining with 4,6-diamidino-2-phenylindole and septa staining with calcofluor:
The protocols described in Gomez and Forsburg (2004) were used for 4,6-diamidino-2-phenylindole (DAPI) and calcofluor staining. For asci staining, h+ and h− strains were mated on SPA plates at 25° and ethanol fixed after 24 hr. Cells were visualized with a Leica DMR microscope. Images were captured with a Hamamatsu (Bridgewater, NJ) digital camera and Improvision (Lexington, MA) Openlab software.
RESULTS
Rationale:
Our screen was based on previous experiments indicating that genes known to be required for S-phase progression show defects in rereplication: the dre phenotype (Fisher and Nurse 1996; Snaith and Forsburg 1999). We used a strain carrying nmt-cdc13+, which expresses cyclin B under a thiamine-repressible promoter. This strain is viable on minimal media, but rereplicates in minimal media plus thiamine or on rich (YES) media up to DNA contents of 8, 16, or even 32C, a lethal phenotype (Hayles et al. 1994; Fisher and Nurse 1996). We expected to isolate mutants with defects in the process of rereplication, which include not only specific S-phase genes but also mutants that might be defective in transcriptional repression of the nmt promoter or degradation of the Cdc13 protein. We used two broad approaches to isolate dre mutants. First, we used a selection for mutants that maintained viability under rereplicating (plus thiamine) conditions (Figure 1A). Independently, we isolated temperature-sensitive mutants in the nmt-cdc13+ strain and screened them individually for defects in rereplication (Figure 2).
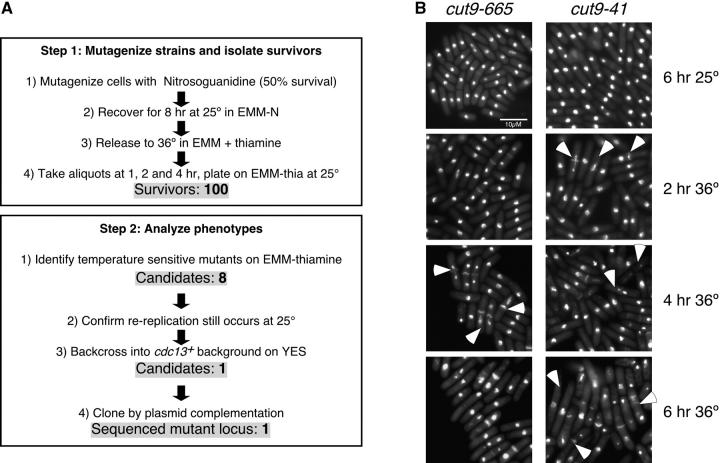
Figure 1.—
(A) The steps and results of the enrichment method approach. (B) Phenotypic comparison of the new cut9-41 allele and cut9-665. Strains cut9-41 (FY1304) and cut9-665 (FY1305) were grown at 25° to OD595 = 0.4 and shifted to 36°. Samples were collected every 2 hr and fixed for DAPI/calcoflour. Arrowheads indicate cells with chromosome and/or septation defects. Bar, 10 μm.
Enrichment selection method—isolation of cut9-41:
Rereplication to high levels is lethal (Moreno and Nurse 1994). We reasoned that mutants that do not rereplicate would be more likely to remain viable; therefore, inducing rereplication should enrich the survivors for mutants that specifically block DNA rereplication. We anticipated that genes required for rereplication might be essential for viability, and therefore any mutant alleles would have to be conditional, so we used high temperature to inactivate any candidate genes. nmt-cdc13+ cells were mutagenized with nitrosoguanidine, allowed to recover for 8 hr at 25°, and then blocked in G1 by nitrogen starvation. Cells were released to 36° in medium plus thiamine to induce rereplication simultaneous with inactivating any candidate genes. Aliquots were harvested at 1, 2, and 4 hr, plated on thiamine-free medium, and incubated at 25°. After 5 days, a total of 100 survivors were recovered. No survivors were obtained from unmutagenized controls. Only eight candidates were both temperature sensitive and still competent for rereplication at 25° in the presence of thiamine. The eight candidates were backcrossed to a wild-type strain to separate the ts mutation from the rereplicating nmt-cdc13 allele; thus, these strains no longer require growth on minimal medium. Seven of the eight mutants were no longer ts when grown on YES, suggesting they had a mutation in some metabolic pathway that inhibited their growth on minimal media at the restrictive temperature, and they were discarded.
The only candidate left was complemented by a genomic clone that expressed the cut9+ gene, a component of the APC. Linkage analysis showed that our candidate was linked to the cut9-665 allele (Samejima and Yanagida 1994; our FY1068), and sequencing of the cut9 gene in the rereplication candidate strain showed that thymidine 1042 was mutated to cytidine, changing serine 348 to proline on the fourth tetratricopeptide repeat (Table 2, Figure 1B). To compare the new cut9 ts allele, cut9-41, to the already characterized cut9-665, we performed a synchronous shift to 36° and collected samples every 2 hr. Cells were fixed and DAPI/calcofluor stained (Figure 1B). Interestingly, after just 2 hr at 36°, cells with the typical cut phenotype and misegregated DNA were observed in cut9-41 but not in the cut9-665 cells (Figure 1B, arrowheads, 2 hr at 36°). By 4 hr at 36°, cut cells and cells with misegregated DNA were observed in both cut9 ts alleles. After 6 hr at 36°, cut9-41 cells had elongated and exhibited septation defects and uneven DAPI-stained bodies. These results indicate that this new isolated temperature-sensitive allele of cut9 has a more severe phenotype than that of the previously characterized strain (Samejima and Yanagida 1994).
TABLE 2.
Genes able to suppress the temperature sensitivity of somedre mutants
| Mutant strain | Genes in recovered plasmid | Known or predicted protein function | Protein mutation |
|---|---|---|---|
| cut9-41 | cut9 (FL+PP) | APC subunit | S 348 P |
| (dre1-41) | SPAC6F12.15c | ||
| dre2-4 | SPA19A8.02 (last 1380 bp) | Hypothetical protein | ND |
| sec73 (FL + PP) | Intracellular protein transport (predicted) | ND | |
| SPAC19A8.01c | |||
| ini1 (last 342 bp) | Involved in mRNA splicing | ND | |
| SPAC23H3.02c | |||
| rad4-42 | rad4 (first 1564 bp + PP) | DNA replication protein | T 45 A |
| (dre3-42) | SPAC23C4.18c | ||
| dre4-54 | hgp1 (FL + PP) | Hyphal growth protein I | W 117 Stop |
| SPAC13C5.02 | |||
| dre6-82 | nuc1/rpa1 (FL + PP) | Large subunit RNA polymerase I | ND |
| SPBC4C3.05c | |||
| sep1 (last 738 bp) | Transcription factor involved in septation | ND | |
| SPBC4C3.12 | |||
| dre7-125 | SPAC1B1.03c (FL + PP) | Nucleocytoplasmatic transport (predicted) | ND |
| SPAC1B1.02c (first 1257 bp + PP) | NAD kinase (predicted) | ND | |
| dre9-141 | snu66 (last 1067 bp) | U4/U6.U5 snRNP component (predicted) | ND |
| SPAC167.03c | |||
| ptb1 (FL + PP) | Geranylgeranyltransferase β | ND | |
| SPAC167.02 | |||
| SPAC167.01 (last 1408 bp) | Unfolded protein response (predicted) | ND | |
|
psf2-209 (dre13-209) |
SPBC725.13c (FL + PP) | Psf2 homolog/DNA replication protein (predicted) | R 133 K |
| dre18-63 | SPAC22H10.05c (FL + PP) | Polyadenylation factor (predicted) | ND |
| SPAC22H10.06c (FL + PP) | Very hypothetical protein | ND | |
| zym1 (FL + PP) | Zinc homeostasis | ND | |
| SPAC22H10.13 | |||
| SPAC22H10.04 (last 333 bp) | Ser/Thr protein phosphatase (predicted) | ND | |
| dre24-8 | SPAC19E9.01c (last 770 bp) | Karyopherin docking complex (predicted) | ND |
| SPAC6F12.17 (FL + PP) | mRNA 3′ end maturation (predicted) | ND | |
| SPAC6F12.16c (first 584 bp + PP) | mRNA helicase involved in mRNA export (predicted) | ND |
dre strains, the mutated gene of which was identified, are underlined. Systematic names of all genes are included. FL, full-length gene; PP, proximal promoter; ND, not determined.
Screening method approach:
The enrichment selection method was not very successful as only one mutant was isolated. Hence, we performed a screen (Figure 2A) based on the prediction that most S-phase genes are likely to be essential. We isolated a bank of temperature-sensitive mutants in strain FY875 (nmt-cdc13+) (Figure 2A, step 1) and screened for their ability to rereplicate when cdc13+ expression was turned off by the addition of thiamine (Figure 2A, step 2). To identify rereplication mutant candidates, first we analyzed the flow cytometry profiles of our rereplication parent strain as shown in Figure 2B (left). FY875 was arrested in G1 and released into S phase in plus-thiamine medium to induce rereplication and incubated at 25° and 36°. By 10 hr at 25°, strain FY875 had a majority of cells with a 4C DNA content, some with a 2C, and very few with an 8C. A similar flow cytometry profile was obtained when cells were incubated for 5 hr at 36°, since cells cycle faster at a higher temperature (Figure 2B, left, shaded background profiles). To identify rereplication mutant candidates, we compared the flow cytometry profiles obtained for the temperature-sensitive mutants at these two time points and temperatures. All those mutants that showed different flow cytometry profiles when comparing 10 hr at 25° vs. 5 hr at 36° were kept for further analyses. Figure 2B (right) shows an example of a rereplication mutant candidate, dre24-8. We required candidates to be proficient for rereplication at 25°, as determined by a 4C DNA content at 10 hr. Mutants that were unable to enter S phase after 5 hr at 36° or were notably delayed compared to its 10-hr profile at 25° were chosen for further analysis.
Figure 2A summarizes the results of the screen. A total of 366 temperature-sensitive mutants were isolated, of which 339 were analyzed by flow cytometry for rereplication defects following release from G1. The remaining 27 ts mutants either were too sick to propagate or were unable to arrest in G1 after nitrogen starvation. Of the candidates that we screened, 42 had rereplication defects. Seventeen were haploid but 25 had diploidized at some point during propagation, probably due to the nmt-cdc13+ background, which tends to accumulate homozygous diploids. To enable further genetic analysis, we attempted to reisolate haploids either by using m-FPA to promote chromosome loss or by tetraploid crosses (see materials and methods). We were successful in haploidizing 7 additional strains, leaving us with 24 haploid candidates for further analysis. These were backcrossed to wild-type strain FY255 to isolate the ts mutation from the nmt-cdc13+ background and to ensure that the mutant phenotype was due to a single locus.
Phenotype characterization:
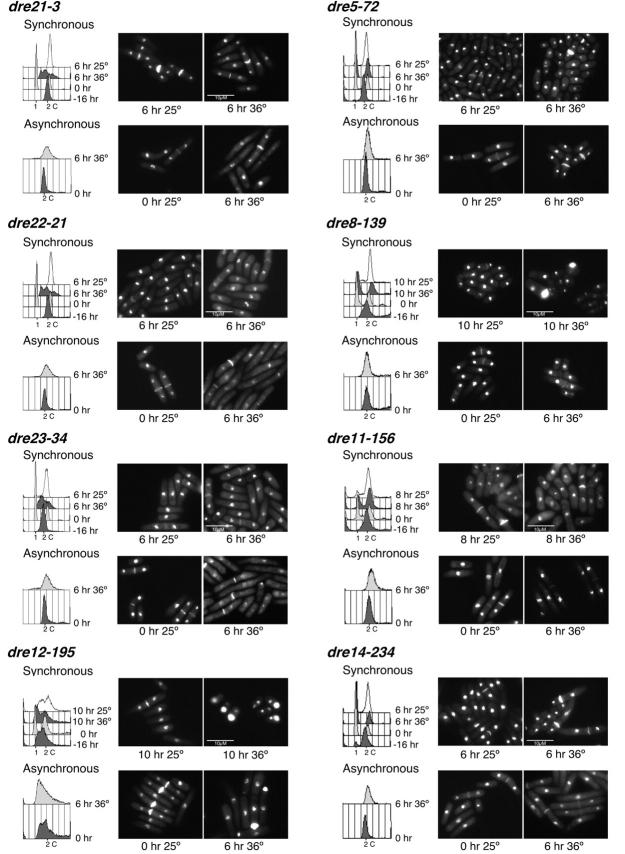
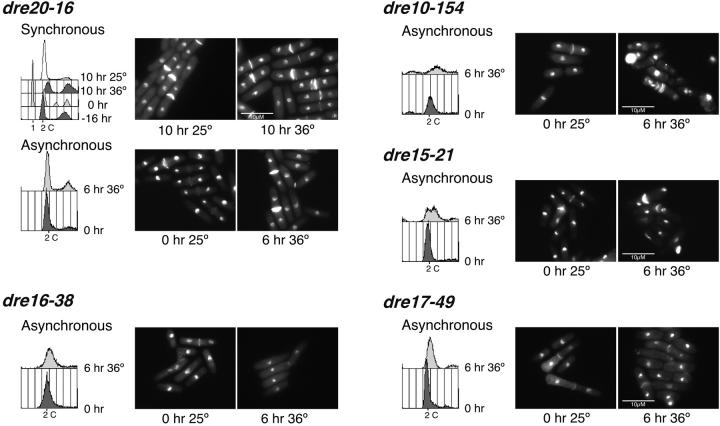
We examined S-phase phenotypes of the mutants in a cdc13+ background following synchronous release from nitrogen starvation (G1 arrest) to the restrictive temperature (Figure 3). Flow cytometry analyses showed that most mutants had defects in S-phase entry, with a substantial fraction of cells remaining with a 1C DNA content even after 6–8 hr. Others showed intermediate DNA contents. Morphological phenotypes were varied, and most were mixed without a single distinct morphology. A few showed a high fraction of cdc or cut cells and a surprising number arrested with a large fraction of septated binucleate cells.
Figure 3.—
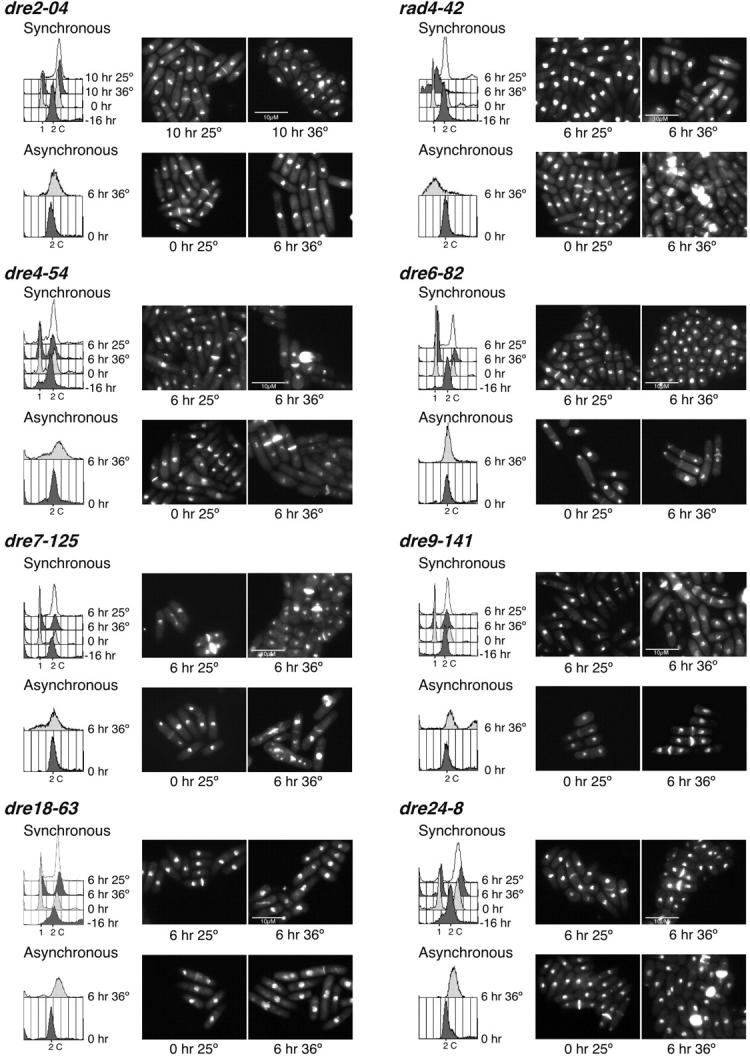
Phenotype of the candidate mutants after synchronous and asynchronous shift to the restrictive temperature. For the synchronous shift, the indicated strains were arrested in G1 by nitrogen starvation and released to 25° and 36°. For the asynchronous shift analysis, cells were grown at 25° to OD595 = 0.4 and shifted to 25° and 36°. Samples were collected every 2 hr, ethanol fixed, and analyzed by flow cytometry and DAPI/calcoflour stained. Six, 8, or 10 hr after the shift to 36° is shown. Some mutants did not arrest in G1 after nitrogen starvation making the synchronous shift analysis impossible. Hence, only their asynchronous shift data are shown.
We also shifted asynchronous, exponentially growing cells to the restrictive temperature. In this case, most mutants arrested with a 2C DNA content, which is typical of many S-phase mutants (e.g., Nasmyth and Nurse 1981). Morphologies were generally similar to those observed for the synchronous shift. These results correlate with the rereplication defects observed when they were first isolated, indicating that, as previously shown (Snaith and Forsburg 1999), a mutant that is unable to rereplicate will also have defects in a normal S phase.
Linkage analysis:
We used classical linkage analysis to determine how many loci are represented in this collection. Linkage analysis is preferred because fission yeast does not form stable diploids required for complementation. As S. pombe is easily manipulated by random spore analysis, we crossed the isolated rereplication mutants to each other and analyzed the percentage of wild-type progeny to determine the frequency of recombinants. If two ts loci are unlinked, we expect to see ∼25% wild-type colonies in the offspring, while allelic mutants will generate very few, if any, wild-type recombinants. This analysis showed that mutants dre21-3, dre22-21, and dre23-34 belong to the same linkage group. This result correlates with their very similar flow cytometry profiles and cell phenotypes observed in the synchronous and asynchronous shift analyses (Figure 3), suggesting that the same gene is mutated. The remaining linkage groups have only one member, indicating that our screen is far from saturating.
Identification of genes:
To identify the cognate genes, we transformed the temperature-sensitive mutants with a genomic DNA library and screened for complementation of the growth defect at 36°. We were able to isolate transformants for 21 mutants. Following incubation at restrictive temperature, we identified transformants that complemented the growth defect at 36° in nine of the strains from which we recovered plasmids. No complementing plasmids were identified in the remaining candidates. For several of the strains, multiple plasmids rescued the temperature growth defect, but in all cases the inserts included overlapping genomic regions. By comparing the different rescuing fragments, a minimum complementing sequence was determined (Table 2).
Table 2 shows the mutant strains, genes present in the minimum fragment that rescued each mutant, the length of each gene and the presence of a proximal promoter, predicted or known function of each encoded protein(s), and, where determined, the amino acid substitution in the protein at the mutated genomic loci. The genes present in the recovered plasmids could correspond to the actual mutated genes or to high-copy suppressors. On the basis of the proposed function of the genes established by curation of the genome, the majority of these genes encode for proteins that, if mutated, could cause indirect defects in DNA replication: for example, RNA metabolism or protein trafficking. We therefore restricted further analysis to candidates that either corresponded to known replication genes or had no known function based on sequence homology and could be novel factors. The identity of the mutant gene was confirmed by linkage analysis and/or by sequencing the genomic locus to confirm the presence of a mutation.
dre3-42 is an allele of rad4/cut5:
dre3-42 was rescued by a plasmid that expressed only the first 1564 bp of the DNA replication gene rad4+ (also called cut5+), truncating the protein at amino acid 480. Therefore, we crossed dre3-42 to the temperature-sensitive strain rad4-116 (our FY1114; Duck et al. 1976) and found that the mutations were linked. Sequencing of the rad4 gene in dre3-42 showed that threonine 45 was substituted for alanine where codon ACG was changed to GCG. Interestingly, the same amino acid is mutated in rad4-116 and cut5-580, but, in both of those cases, substituted by methionine. As dre3-42 is a new temperature-sensitive allele of rad4, we will name it rad4-42 from here on. The phenotype of rad4-42 was identical to that observed for previous alleles, with a high fraction of cut cells and less than G1 DNA content as seen by flow cytometry analysis (Figure 3). Consistent with this, a previous study (Snaith and Forsburg 1999) showed that rad4-116 had rereplication defects when cells were induced to rereplicate by overexpression of the Rum1 inhibitor.
dre4+ encodes a WW domain protein:
The dre4-54 rescuing plasmid had two genes (Table 2). One encoded protein SPAC13C5.02; the other was nuclear fusion protein 1, Tht1 (Tange et al. 1998). Because the plasmid contained only part of tht1+, we reasoned that the most feasible candidate was the former gene. We subcloned wild-type SPAC13C5.02 and verified that it could rescue dre4-54 temperature sensitivity. To confirm that the gene was actually mutated, we amplified and sequenced dre4-54 and found that codon 117 TGG was changed to TAG, generating a premature stop codon (Table 2). Thus, dre4-54 corresponds to SPAC13C5.02. Dre4 has a WW domain and an FF domain, both protein-binding motifs. WW domains are common in diverse proteins, often in multiple copies, and are thought to bind proline-rich ligands. They may be regulated by tyrosine phosphorylation (Sudol et al. 2001; Ilsley et al. 2002). The FF domain is thought to be a phosphopeptide-binding motif and frequently accompanies WW domains (Bedford and Leder 1999; Allen et al. 2002).
Synchronous and asynchronous shift analyses showed that dre4-54 has a heterogenous phenotype and suggest that this ts allele is not completely penetrant or that the gene has multiple functions. A fraction of cells block as septated binucleates. More strikingly, the nuclear structure of many cells is abnormal and the chromatin appears hypercondensed. Some cells have aberrant calcofluor staining, suggesting delocalized septal material. Cells with misegregated DNA are also observed. Flow cytometry analysis suggests that the binucleate cells have a 2C DNA content, consistent with each nucleus being arrested in G1. Interestingly, a small fraction of cells with less than 2C DNA content appear when dre4-54 cells are grown asynchronously, and this number is increased after shifting the cells for 6 hr at 36°.
Psf2 homolog characterization—identification of psf2-209:
The minimal fragment that rescued dre13-209 had genes that encoded two different proteins, an acetyl glutamate synthase and a hypothetical protein related to S. cerevisiae and Xenopus psf2+ (partner of Sld five 2). Psf2p is part of a novel replication complex, GINS, which was recently shown to be essential for initiation and elongation of DNA replication in budding yeast and Xenopus (Kanemaki et al. 2003; Kubota et al. 2003; Takayama et al. 2003). As this gene was the best candidate, we verified that dre13-209 was rescued with a plasmid expressing psf2+ alone (kindly provided by H.-K. Huang).
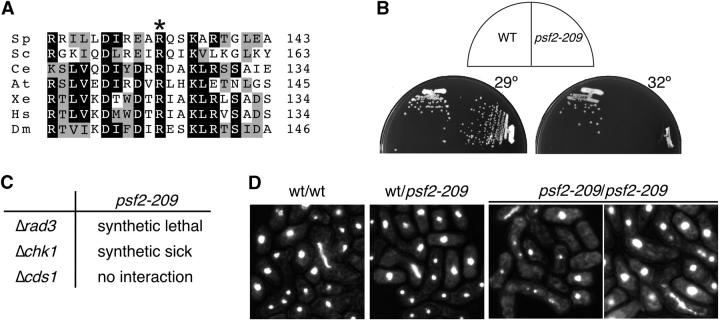
To confirm that the gene was actually mutated, we amplified and sequenced dre13-209 psf2 and found that codon 133 AGA (R) was changed to AAA (K) (Table 2). These results demonstrate that dre13-209 is a temperature-sensitive allele of psf2; thus, we named it psf2-209. Interestingly, the arginine residue mutated in this allele is conserved in all psf2 homologs identified, from budding yeast to humans and plants (Figure 4A). S. pombe Psf2 is 34% identical and 51% similar to S. cerevisiae PSF2 and 33% identical and 53% similar to the Xenopus homolog. Similar values are observed for the Caenorhabditis elegans, Arabidopsis, human, or Drosophila proteins (see alignment in Takayama et al. 2003). psf2-209 mutant cells grow as wild type at 25° and 29° but at 32° and higher temperatures they are unable to form colonies (Figure 4B).
Figure 4.—
(A) Partial amino acid sequence alignment of S. pombe (Sp) Psf2 and their homologs from S. cerevisiae (Sc), C. elegans (Ce), Arabidopsis thaliana (At), Xenopus (Xe), Homo sapiens (Hs), and Drosophila melanogaster (Dm). The Psf2 sequences were aligned with the MAP Multiple Sequence Alignment program. White letters on a black background indicate that identical amino acids are present in four or more Psf2 proteins. Black letters on a gray background indicate similar amino acids present in four or more Psf2 proteins. Numbers to the right indicate the last amino acid shown in this comparison. The conserved arginine, mutated to lysine in S. pombe Psf2 temperature-sensitive protein, is marked with an asterisk. (B) psf2-209 temperature sensitivity. S. pombe wild-type (FY255) and psf2-209 (FY2712) strains were streaked onto YES plates and incubated at the indicated temperatures. (C) Synthetic interactions between psf2-209 and damage checkpoint genes. psf2-209 (FY2711) was mated to strains Δrad3 (FY1107), Δchk1 (FY421), and Δcds1 (FY865) on SPA and allowed to sporulate. Diploids were patched on YES and spores analyzed by tetrad analysis and replica plating to EMM− ura. (D) psf2-209 diploids have meiotic defects. Wild-type and psf2-209 haploids (FY254, FY255, FY2711, and FY2712) were mated on SPA plates for 24 hr, ethanol fixed, and DAPI stained.
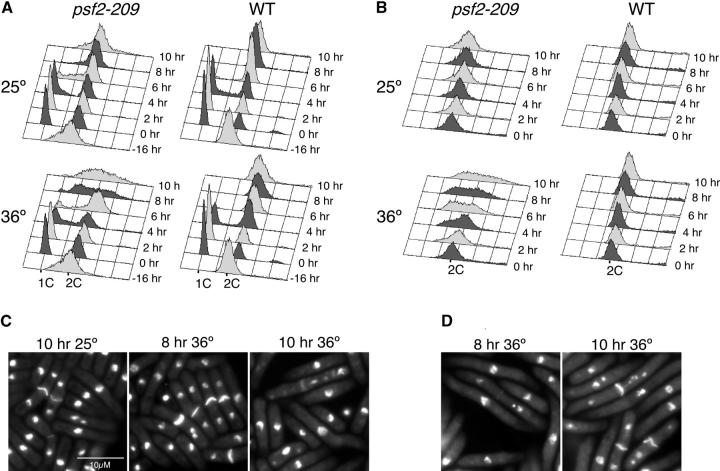
Next, we analyzed psf2-209 cells shifted to the restrictive temperature of 36°. When cells were arrested in G1 by nitrogen starvation and released to 36°, flow cytometry analysis showed that by 6 hr the majority of the cells had a 2C DNA content, indicating that they had completed bulk DNA synthesis (Figure 5A). Interestingly, after 8 and 10 hr at 36° the flow cytometry peak broadened, showing cells with DNA contents between 1C and 2C. This result suggests that psf2-209 mutants arrest in the second cell cycle. DAPI and calcofluor staining analysis show cells with misegregated nuclei after 8 hr at 36° and, by 10 hr, elongated cells, some of which had more than two DAPI-stained bodies (Figure 5B).
Figure 5.—
psf2-209 has a heterogenous phenotype. Wild-type (FY254) and psf2-209 (FY2712) strains were blocked in G1 by nitrogen starvation and released to 25° or 36° (synchronous shift, A and B) or grown at 25° to OD595 = 0.4 and shifted to 25° and 36° (asynchronous shift, C and D). Samples were collected every 2 hr, ethanol fixed, and analyzed by flow cytometry (A and C) and DAPI/calcoflour stained (B and D). Bar, 10 μm.
When asynchronously growing cells were shifted to 36°, flow cytometry profiles showed that cells had DNA contents between 1C and 2C at 4 hr (Figure 5C). After 6–10 hr at 36°, profiles had broadened and looked like the 8 and 10 hr of the synchronous shift to 36° (Figure 5, A and C). DAPI/calcofluor staining and microscopy analysis of psf2-209 showed a high fraction of cells with an elongated cdc phenotype, others with more than two DAPI-stained bodies, and some with uneven segregation of their DNA (Figure 5D). Compared to wild-type cells (FY254), psf2-209 mutants show a delayed entry and/or slower S-phase progression even at the permissive temperature (Figure 5A; compare the 4- and 6-hr profiles).
psf2 ts genetically interacts with damage checkpoint mutants:
Many replication mutants cause DNA damage that activates replication checkpoints; in the absence of the checkpoints, the cells may lose viability or change phenotype at the restrictive temperature. We crossed psf2-209 to the checkpoint deletion strains rad3 (FY1107), chk1 (FY421), and cds1 (FY865). Rad3p (ATM/ATR homolog) is required for the replication and damage checkpoints and is thought to act upstream of Chk1p and Cds1p; Chk1p responds mainly to the DNA damage checkpoint, while Cds1p responds to the DNA replication checkpoint (Huberman 1999; Rhind and Russell 2000). We could not recover a psf2-209 Δrad3 double mutant even at the permissive temperature. The double mutant with Δchk1 had a slow growth phenotype compared with the single mutants and formed microcolonies. No genetic interaction was observed with Δcds1 (Figure 4C). These results suggest that the replication damage checkpoint pathway is required to maintain normal viability of psf2-209 cells even at the permissive temperature, suggesting that the mutant has DNA damage even at the permissive temperature.
psf2-209 homozygous diploids have meiotic defects:
In the course of our analysis, we examined meiotic progression in asci resulting from mating h+ and h− psf2-209 haploids on a sporulation plate at the permissive temperature. Haploids were crossed and ethanol fixed after 24 hr. Zygotes were analyzed by DAPI staining and DIC/Nomarski microscopy. Nearly all homozygote wild-type asci showed normal horse tails, two (meiosis I) or four (meiosis II) DAPI-stained bodies of equal size, and normal spore shape. The homozygous psf2-209/psf2-209 asci showed aberrant horse tail structures, unequal DAPI-stained bodies, and when present, spores of different sizes (Figure 4D). These results suggest that Psf2p is essential for the normal progression of meiosis, although they do not indicate which stage of the process is disrupted.
DISCUSSION
The screen performed in this article was designed to identify temperature-sensitive mutants defective in the process of replication regardless of morphology by isolating strains unable to rereplicate their DNA in the absence of Cdc13p cyclin. While this screen succeeded in isolating many new mutants, in addition to several new alleles of known genes, the rate of return for specific S-phase genes was relatively low, given the number of mutants originally isolated. Approximately 10% of the ts mutants analyzed had some rereplication defect, which was considerably higher than we would have expected. This suggests that the rereplication phenotype that we used as a basis for selection is extremely sensitive to general cell growth defects. This was confirmed by the fact that the majority of the genes present in the rescuing plasmids encoded for proteins involved in RNA metabolism or protein trafficking.
Unfortunately, we were unable to rescue plasmids from 14 interesting temperature-sensitive haploid mutants. A different genomic or cDNA library or perhaps a different cloning approach could be used to identify the mutated genes in these strains. Furthermore, >40% of the isolated ts mutants with rereplication defects were homozygous diploids, and we were unable to haploidize them to continue with their formal genetic analysis. Both haploidization methods used, m-FPA and tetraploidization, were equally inefficient. In addition to these two methods, a third approach was employed with no success. We used a plasmid expressing mat1-P, pON104 (kindly provided by Olaf Neilsen); no colonies were obtained after transformation, possibly because cells expressing the plasmid were induced to sporulate, causing cell death (data not shown).
Although we have not been able to identify the mutated genes corresponding to the majority of the isolated temperature-sensitive strains, we did partially characterize their ts phenotype. We performed synchronous and/or asynchronous temperature shifts of the 24 temperature-sensitive haploid strains isolated, followed by DAPI/calcoflour staining and flow cytometry analysis, with results consistent with defects in replication. Mapping and cloning efforts continue in our laboratory.
Among the genes that we succeeded in identifying, we isolated a novel temperature-sensitive allele of cut9+, cut9-42. Cut9p is a component of the APC, and its role in mitosis is well established (Samejima and Yanagida 1994; Yamada et al. 1997). Because the APC promotes the degradation of the mitotic cyclins Cdc13p and Cig1p (Kominami et al. 1998; Blanco et al. 2000), we posit that the cut9 mutant affects rereplication because these cells cannot degrade the Cdc13p cyclin appropriately in the presence of thiamine. Thus, the survival of the cut9-41 mutant might be an artifact of the screening approach. Interestingly, this allele is more penetrant than cut9-665 and could be used to further characterize the various roles of the anaphase-promoting complex.
We also isolated a new temperature-sensitive allele of rad4+, a known fission yeast gene involved in DNA replication and checkpoint control. Interestingly, our ts allele, rad4-42, has a mutation in the same amino acid as the previously characterized rad4 alleles, rad4-114 and cut5-580 (Hirano et al. 1986; Saka et al. 1997), but instead of threonine 45 changing to methionine as in the previous cases, it changed to alanine. This result suggests that this residue tolerates different amino acid changes making the protein temperature sensitive. A C-terminal truncated gene rescued the temperature sensitivity of rad4-42, showing that the last 168 amino acids of the Rad4p are not essential for its function. This agrees with previous data that the C-terminal region of rad4 is nonessential (Fenech et al. 1991; Saka and Yanagida 1993).
We also isolated a novel temperature-sensitive mutant, dre4-54, which is defective in a gene containing an uncharacterized WW domain protein. Our ts mutant has codon 117 changed to a stop codon, the same mutation obtained by H. C. Joshi who suggested the name hgp1+ for hyphal growth phenotype (H. C. Joshi, personal communication). However, we see no resemblance to a hyphal or pseudohyphal phenotype. A fraction of the cells appear to arrest as binucleates with a septum, but unseparated phenotypes are observed for a diverse number of cell cycle mutants, including cytokinesis mutants (Gould and Simanis 1997) and the guanine nucleotide exchange factor pim1-d1 (Demeter et al. 1995), and may be evidence of the coupling between cytokinesis and G1. Our flow cytometry analysis suggests that the nuclei of the unseptated cells have arrested in G1 since a 2C peak is detected after shifting the cells to 36°. More strikingly, the dre4 mutant has a disordered nuclear structure, looks hypercondensed, and suffers abnormal chromosome segregation. Again, this is reminiscent of a variety of other strains, including pim1-d1 (Demeter et al. 1995) and topoisomerase mutants (Uemura and Yanagida 1984). Whether dre4+ affects DNA synthesis directly or its rereplication phenotype is an indirect result of other defects in nuclear structure or maintenance remains to be determined. The closest relatives to Dre4 using a BLAST search are uncharacterized ORFs found in other fungi—in Magnaporthe (accession no. EAA57360) and Gibberella (accession no. XP_385543; E values ≤ 2e × 10−22; data not shown)—although there are related proteins in many species.
The most interesting temperature-sensitive mutant identified in this screen is psf2-209. Homologs of psf2+ have been recently identified and characterized in S. cerevisiae and Xenopus as a component of the novel replication complex GINS (Kanemaki et al. 2003; Kubota et al. 2003; Takayama et al. 2003). Our allele is the first temperature-sensitive GINS mutant in fission yeast, and its further analysis will help to elucidate the role of the GINS complex. psf2-209 cells at 36° can replicate their DNA and arrest with a 2C DNA content. A similar DNA content is seen with the minichromosome maintenance replication proteins, which ts mutants also arrest with a 2C DNA content (Coxon et al. 1992; Miyake et al. 1993; Forsburg and Nurse 1994; Takahashi et al. 1994; Liang and Forsburg 2001). However, unlike mcm mutants, psf2-209 arrested cells have a mixed phenotype with some cells elongated, others with misegregated DNA with or without a septum, and others with more than two DAPI-stained bodies, suggesting either fragmentated DNA or the presence of lagging chromosomes. Importantly, this is not the typical premature mitosis phenotype observed in other replication mutants that block cells prior to initiation of DNA synthesis, including rad4 (Fenech et al. 1991), orp1 (Grallert and Nurse 1996), cdc18 (Kelly et al. 1993), and pol1 (D'Urso et al. 1995) mutants, because psf2-209 cells synthesize DNA (as seen by flow cytometry).
The misegregated DNA phenotype observed for psf2-209 cells at the restrictive temperature suggests that Psf2p might have more than one function. Interestingly, psf2+ was recently identified as a high-copy suppressor of a temperature-sensitive allele of the passenger protein Bir1p. Furthermore, this group showed that Psf2p is needed for the proper localization of Bir1p linking psf2+ to chromosome segregation (H.-K. Huang, J. M. Bailis, E. B. Gómez, J. Leverson, S. L. Forsburg and T. Hunter, unpublished results).
Strikingly, psf2-209 cells mate at the permissive temperature but go through an aberrant meiosis. It will be very interesting to examine whether this phenotype is related to its replication or to chromosome segregation function. When analyzing other nonreplicating mutants, we have observed that mutants with no apparent chromosome segregation defects in vegetative growth have a high percentage of asci going through an aberrant meiosis (E. B. Gómez and S. L. Fosrburg, unpublished results). This would suggest that the meiotic cells are much more sensitive to DNA segregation defects than are vegetatively growing cells.
Our screen was successful in identifying several interesting new genes, but proved difficult and time consuming in its execution. Many of the mutants that we isolated are refractory to transformation and cloning. Additionally, it is clear that the screen was not saturating, since only one previously know replication gene (rad4+) was isolated. Since our methodology relied upon the isolation of a bank of temperature-sensitive strains and then a labor-intensive screening protocol, it is not surprising that we missed many genes. Moreover, a range of interesting genes that either are unlikely to produce ts alleles or are nonessential for growth were certainly overlooked. Thus, there is still a place for a nonbiased genetic screen for replication mutants in fission yeast.
Acknowledgments
We thank Sebastian Laría, Irma Padilla, Ciana Palencia, Lisa Scott, and Rion Snow for assistance in mutant isolation and characterization. Thanks go to Harish Joshi, Han-Kuei Huang, and Tony Hunter for communication of results prior to publication, to Han-Kuei Huang and Tony Hunter for the psf2+ plasmid, and to Tony Carr for the fission yeast genomic library. We are grateful to William Dolan and Julie Bailis for helpful comments on the manuscript. We thank Lorraine Pillus for her hospitality to E.B.G. during preparation of this manuscript. This work was supported by grants to S.L.F. from the National Science Foundation (MCB 9974732) and the National Institutes of Health (GM-059321).
References
- Allen, M., A. Friedler, O. Schon and M. Bycroft, 2002. The structure of an FF domain from human HYPA/FBP11. J. Mol. Biol. 323: 411–416. [DOI] [PubMed] [Google Scholar]
- Bedford, M. T., and P. Leder, 1999. The FF domain: a novel motif that often accompanies WW domains. Trends Biochem. Sci. 24: 264–265. [DOI] [PubMed] [Google Scholar]
- Blanco, M. A., A. Sanchez-Diaz, J. M. de Prada and S. Moreno, 2000. APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J. 19: 3945–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek, D., R. Bartlett, K. Crawford and P. Nurse, 1991. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature 349: 388–393. [DOI] [PubMed] [Google Scholar]
- Coxon, A., K. Maundrell and S. E. Kearsey, 1992. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 20: 5571–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter, J., M. Morphew and S. Sazer, 1995. A mutation in the RCC1-related protein pim1 results in nuclear envelope fragmentation in fission yeast. Proc. Natl. Acad. Sci. USA 92: 1436–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duck, P., A. Nasim and A. P. James, 1976. Temperature-sensitive mutant of Schizosaccharomyces pombe exhibiting enhanced radiation sensitivity. J. Bacteriol. 128: 536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Urso, G., B. Grallert and P. Nurse, 1995. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J. Cell Sci. 108(Pt. 9): 3109–3118. [DOI] [PubMed] [Google Scholar]
- Fenech, M., A. M. Carr, J. Murray, F. Z. Watts and A. R. Lehmann, 1991. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe: a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res. 19: 6737–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D. L., and P. Nurse, 1996. A single fission yeast mitotic cyclin B p34(Cdc2) kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 15: 850–860. [PMC free article] [PubMed] [Google Scholar]
- Forsburg, S. L., and P. Nurse, 1994. The fission yeast cdc19+ gene encodes a member of the MCM family of replication proteins. J. Cell Sci. 107: 2779–2788. [DOI] [PubMed] [Google Scholar]
- Gomez, E. B., and S. L. Forsburg, 2004. Analysis of the fission yeast Schizosaccharomyces pombe cell cycle. Methods Mol. Biol. 241: 93–111. [DOI] [PubMed] [Google Scholar]
- Gomez, E. B., M. G. Catlett and S. L. Forsburg, 2002. Different phenotypes in vivo are associated with ATPase motif mutations in Schizosaccharomyces pombe minichromosome maintenance proteins. Genetics 160: 1305–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, K. L., and V. Simanis, 1997. The control of septum formation in fission yeast. Genes Dev. 11: 2939–2951. [DOI] [PubMed] [Google Scholar]
- Grallert, B., and P. Nurse, 1996. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 10: 2644–2654. [DOI] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Lopreno, 1974 Schizosaccharomyces pombe. Plenum Press, New York.
- Hayles, J., D. Fisher, A. Woollard and P. Nurse, 1994. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2 mitotic B cyclin complex. Cell 78: 813–822. [DOI] [PubMed] [Google Scholar]
- Hirano, T., S. Funahashi, T. Uemura and M. Yanagida, 1986. Isolation and characterization of Schizosaccharomyces pombe cut mutants that block nuclear division but not cytokinesis. EMBO J. 5: 2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman, J. A., 1999. DNA damage and replication checkpoints in the fission yeast, Schizosaccharomyces pombe. Prog. Nucleic Acid Res. Mol. Biol. 62: 369–395. [DOI] [PubMed] [Google Scholar]
- Ilsley, J. L., M. Sudol and S. J. Winder, 2002. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell. Signal. 14: 183–189. [DOI] [PubMed] [Google Scholar]
- Kanemaki, M., A. Sanchez-Diaz, A. Gambus and K. Labib, 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–724. [DOI] [PubMed] [Google Scholar]
- Kelly, T. J., G. S. Martin, S. L. Forsburg, R. J. Stephen, A. Russo et al., 1993. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74: 371–382. [DOI] [PubMed] [Google Scholar]
- Kohli, J., H. Hottinger, P. Munz, A. Strauss and P. Thuriaux, 1977. Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics 87: 471–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami, K.-I., I. Ochotorena and T. Toda, 1998. Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF(Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells 3: 721–735. [DOI] [PubMed] [Google Scholar]
- Kubota, Y., Y. Takase, Y. Komori, Y. Hashimoto, T. Arata et al., 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 17: 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, D. T., and S. L. Forsburg, 2001. Characterization of Schizosaccharomyces pombe mcm7+ and cdc23+ (MCM10) and interactions with replication checkpoints. Genetics 159: 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake, S., N. Okishio, I. Samejima, Y. Hiraoka, T. Toda et al., 1993. Fission yeast genes nda1+ and nda4+, mutations of which lead to S-phase block, chromatin alteration and Ca2+ suppression, are members of the CDC46/MCM2 family. Mol. Biol. Cell 4: 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, M., and M. Sipiczki, 1993. Polyploidy in the haplontic yeast Saccharomyces pombe: construction and analysis of strains. Curr. Genet. 24: 45–52. [DOI] [PubMed] [Google Scholar]
- Moreno, S., and P. Nurse, 1994. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature 367: 236–242. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and P. Nurse, 1981. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 182: 119–124. [DOI] [PubMed] [Google Scholar]
- Rhind, N., and P. Russell, 2000. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113(Pt 22): 3889–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka, Y., and M. Yanagida, 1993. Fission yeast cut5+, required for S-phase onset and M-phase restraint, is identical to the radiation-damage repair gene rad4+. Cell 74: 383–393. [DOI] [PubMed] [Google Scholar]
- Saka, Y., F. Esashi, T. Matsusaka, S. Mochida and M. Yanagida, 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11: 3387–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima, I., and M. Yanagida, 1994. Bypassing anaphase by fission yeast cut9 mutation: requirement of cut9+ to initiate anaphase. J. Cell Biol. 127: 1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith, H. A., and S. L. Forsburg, 1999. Rereplication phenomenon in fission yeast requires MCM proteins and other S phase genes. Genetics 152: 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol, M., K. Sliwa and T. Russo, 2001. Functions of WW domains in the nucleus. FEBS Lett. 490: 190–195. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., H. Yamada and M. Yanagida, 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell 5: 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, Y., Y. Kamimura, M. Okawa, S. Muramatsu, A. Sugino et al., 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange, Y., T. Horio, M. Shimanuki, D. Q. Ding, Y. Hiraoka et al., 1998. A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J. Cell Biol. 140: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., and M. Yanagida, 1984. Isolation of type I and type II topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and organization. EMBO J. 3: 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, H., K. Kumada and M. Yanagida, 1997. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J. Cell Sci. 110: 1793–1804. [DOI] [PubMed] [Google Scholar]