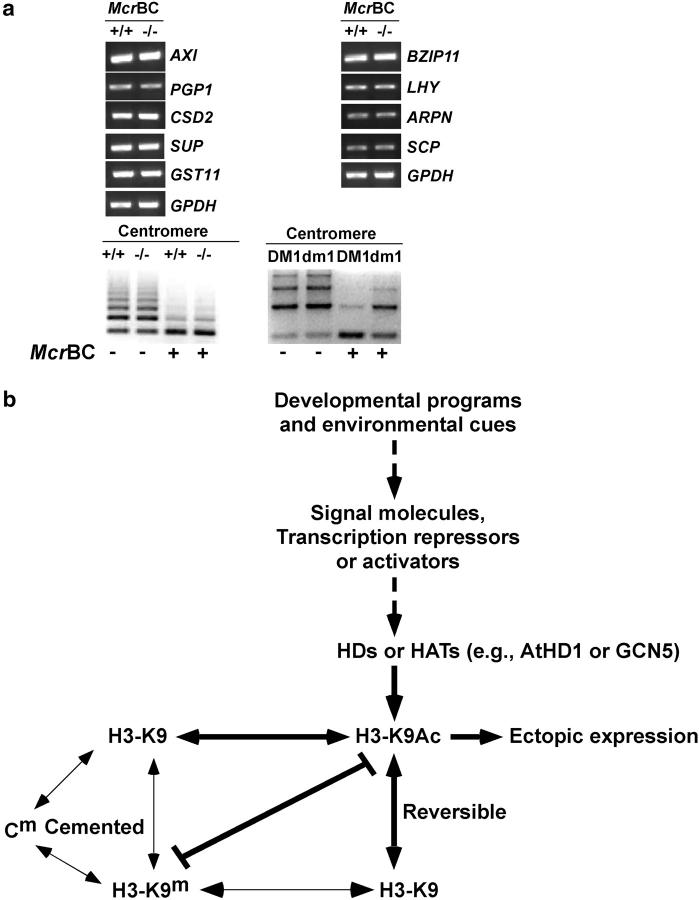
Figure 4.—
DNA methylation in athd1-t1 lines and a model for the role of histone acetylation and deacetylation in plant development. (a) DNA methylation was not affected in genes that were up- or downregulated by histone acetylation or methylation. As a control, centromeric repeats were demethylated in the ddm1 mutant but not in the athd1-t1 line. (b) A simple model indicates that reversible modifications of histone acetylation and deacetylation provide a flexible regulatory system for changes in gene expression in response to developmental programs and environmental cues. The developmental and environmental signals are perceived by signal molecules, transcriptional activators, and repressors that recruit HATs (e.g., GCN5) and HDs (e.g., AtHD1, RPD3), respectively, resulting in changes in histone acetylation or deacetylation (e.g., H3-K9), which lead to transcriptional activation or repression. These changes are reversible when the signals are absent, although changes in histone acetylation/deacetylation are coupled with other cemented changes in histone or DNA methylation as previously reviewed (Jenuwein and Allis 2001; Richards and Elgin 2002), which may induce other epigenetic lesions. Thick, dashed, and thin arrows/lines indicate the interactions that are supported by the results from this report, this and other studies, and previous work, respectively (see text).