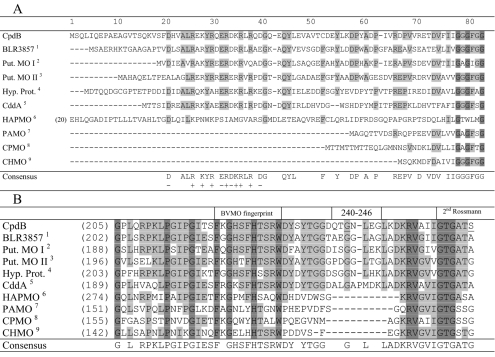
FIG. 2.
(A) Identification of a conserved cluster of charged amino acids in the N-terminal sequences of a subset of BVMO sequences. The highly conserved sequences are shaded. CHMO, CPMO, and PAMO are representative BVMOs that do not have the extraneous N-terminal extension. The FAD-binding motif (GxGxxG, in dark shade) serves as a point of reference. In HAPMO, the first 19 amino acids are not shown; an alternative alignment provided a better FAD-binding motif (25). The origins of the various sequences, besides CpdB (this study), are shown as superscripts in the far left column as follows: 1, B. japonicum (Q89NI1); 2, S. avermitilis (Q82IY8); 3, Streptomyces coelicolor (Q9RL17); 4, M. paratuberculosis (Q73U59); 5, R. ruber (Q938F6); 6, P. fluorescens ACB (AAK54073); 7, T. fusca (1W4X_A); 8, Comamonas sp. strain NCIMB 9872 (Q8GAW0); and 9, Acinetobacter sp. strain NCIMB 9871 (BAA86293). The characters in parentheses are GenBank accession or reference numbers. Put., putative; Hyp. Prot., hypothetical protein. (B) Sequence alignment showing an apparent insertional sequence in CpdB and related sequences in between the BVMO fingerprint and the second Rossmann fold motif, as indicated. Amino acids 240 to 246 in CpdB are the deleted region, and the underlined G and S residues indicate the G242A and S261A substitutions (this study). The references indicated by superscripts are as described in the legend to panel A.