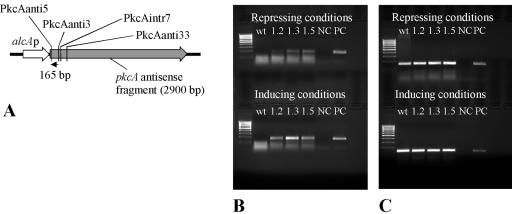
FIG. 5.
Detection of pkcA antisense RNA in A. nidulans transformant strains. (A) Scheme showing the strategy to detect antisense RNA by RT-PCR. The predicted antisense RNA was transcribed to cDNA using the specific primer PkcAanti3. The cDNA is indicated by a black arrow. Primers PkcAanti5 and PkcAanti3 were used in subsequent PCRs, yielding a 165-bp PCR product. The binding sites of the reverse primers PkcAintr7 and PkcAanti33, which were used in additional RT-PCR assays (see the text), are also indicated. (B) Detection of pkcA antisense RNA. RT-PCR was performed using primers specific for antisense RNA (primer PkcAanti3). Cultures for RNA extraction were grown either with glucose (repressing conditions) or with lactose plus 10 mM cyclopentanone (inducing conditions). wt, wild-type strain R21OLAB7; 1.2, 1.3, or 1.5, pkcA antisense-encoding transformant strain PkcAinv1.2, PkcAinv1.3, or PkcAinv1.5, respectively; NC, negative control (no template); PC, positive control (chromosomal DNA as the template). (C) Detection of tef1 RNA. RT-PCR was performed using primers specific for the tef1 gene (primer Tef1rev) as a control. Cultures for RNA extraction were grown with either glucose (repressing conditions) or lactose plus 10 mM cyclopentanone (inducing conditions).