Figure 3.—
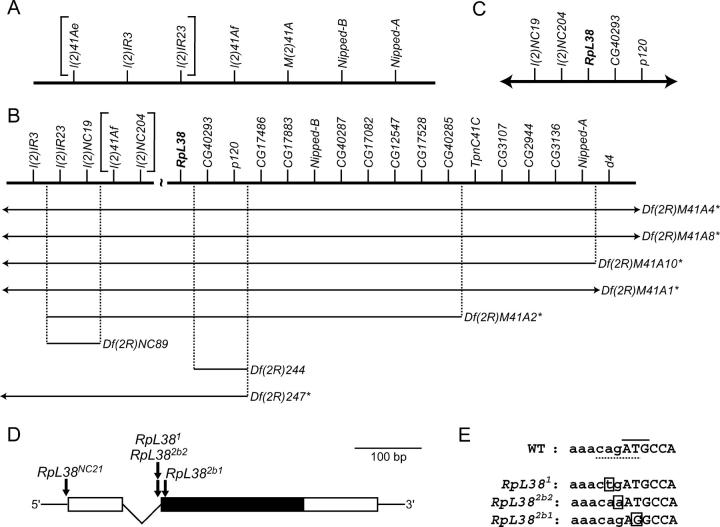
E-2b is RpL38. (A) Genetic map showing the genes in h46 that were identified at the start of this project. The correct ordering of the l(2)41Ae, l(2)IR3, and l(2)IR23 loci (bracketed) was unknown at this time. (B) Map of the h46 region adapted from Corradini et al. (2003) and Myster et al. (2004). RpL38 (boldface type) and genes to the right of RpL38 were positioned on the physical map (not drawn to scale here) whereas genes to the left of RpL38 were ordered on a genetic map. The extent of the gap between the genetic and physical maps was not determined and the correct ordering of the l(2)41Af and l(2)NC204 loci (bracketed) at the distal end of the genetic map was unknown. Note that the l(2)41Ae mutation shown in A is now thought to be a deletion that removes l(2)IR23, l(2)IR3, and five other more proximal genes (Myster et al. 2004). Deficiencies are drawn and annotated as described in the legend to Figure 2. Here, deficiency strains were tested for their ability to complement the E-2b1 and E-2b2 mutations. (C) Physical map (not to scale) of the correct gene order in the vicinity of the RpL38 gene (boldface type). The centromere is to the left and the 2R euchromatin is to the right in A–C. (D) RpL38 gene structure showing the location of mutations. Open boxes, UTRs; solid box, protein-coding region; thin lines, noncoding sequences. (E) DNA sequence surrounding the initiating ATG codon of the RpL38 gene in wild-type (WT) and mutant strains. Lowercase letters, intronic sequence; uppercase letters, protein-coding sequence; solid line, initiating ATG codon; dotted line, splice-acceptor sequence; boxed letter, point mutation.