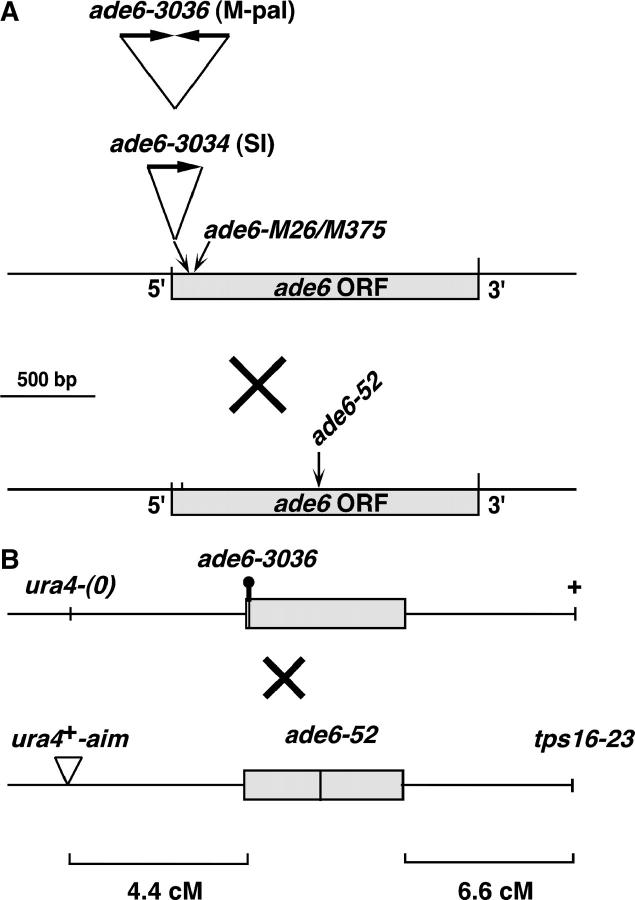
Figure 1.—
Genetic markers used in this study. (A) ade6 alleles. The ade6-3034 and ade6-3036 alleles, described by Farah et al. (2002), consist of one or two copies, respectively, of an 80-bp DNA segment inserted at the BamHI site (bp 24) of the ade6 ORF (GenBank accession no. M37264). The DNA corresponds to 76 bp from the S. cerevisiae mat-a-stk locus (bp 2044–2119 of GenBank accession no. V01313; Ray et al. 1991), plus a 5′ 4-bp BamHI compatible extension. The inserted DNA is not drawn to scale. Both alleles confer an Ade− phenotype. The ade6-M26 allele is a G-to-T transversion at bp 136 of the ade6 ORF and the ade6-M375 allele is a G-to-T transversion at bp 133 of the ade6 ORF (Szankasi et al. 1988). The common test allele ade6-52 is a G-to-A transition at bp 796 of the ade6 ORF (M. E. Fox, unpublished results) and is separated by 773 and 661 bp from ade6-3036 and ade6-M26, respectively. (B) Markers flanking ade6. The ura4+-aim insertion and tps16-23 (temperature-sensitive) markers were described previously (Grimm et al. 1994); tps16 is identical to ags1, 56.1 kb centromere distal to ade6 (Hochstenbach et al. 1998). ura4+-aim is an insertion of a 1.8-kb ura4+ cassette ∼15 kb from ade6. (0) indicates the absence of an insertion. The diagram is not drawn to scale.