TABLE 1.
Estimated parameters of QTL identified in the genome scan for the corn pedigree data
| Single QTL model
|
Multi-QTL model
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| QTL | Linkage group |
Position (cM) |
Flanking markers |
Confidence interval |
σ2u | σ2v | σ2 | ĥ2u (%) | LOD | Variance | Heritability (%) |
| qtl1 | 1 | 55.8 | M098–M188 | 54.8–60.8 | 95.51 | 47.13 | 2.29 | 65.90 | 4.66 | 4.72 | 2.62 |
| qtl2 | 4 | 142.0 | M153–M178 | 140.0–144.0 | 124.74 | 64.18 | 2.30 | 65.23 | 5.05 | 97.18 | 53.85 |
| qtl3 | 5 | 140.9 | M156–M157 | 138.9–146.9 | 93.01 | 58.12 | 3.23 | 60.26 | 3.81 | 7.73 | 4.28 |
| qtl4 | 5 | 175.9 | M076–M007 | 172.9–191.9 | 81.79 | 50.56 | 2.08 | 60.84 | 5.80 | 13.48 | 7.47 |
| qtl5 | 8 | 86.8 | M035–M174 | 86.3–88.2 | 237.09 | 56.52 | 1.49 | 80.34 | 4.84 | 6.14 | 3.40 |
| qtl6 | 9 | 122.9 | M133–M015 | 120.9–126.9 | 99.38 | 52.05 | 1.94 | 64.80 | 5.78 | 16.16 | 8.95 |
| qtl7 | — | M097 | M097 | — | 57.66 | 77.29 | 0.00 | 72.73 | 3.94 | 1.69 | 0.94 |
| qtl8 | — | M028 | M028 | — | 79.29 | 63.41 | 0.90 | 55.22 | 5.44 | 2.81 | 1.56 |
| Polygene | 27.86 | 15.44 | |||||||||
| Residual | 2.69 | 1.49 | |||||||||
σ2u is the estimated genetic variance for QTL, σ2v is the polygenic variance, σ2 is the residual variance, and ĥ2u is the proportion of the total variance contributed by the QTL and is expressed as 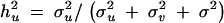 .
.
