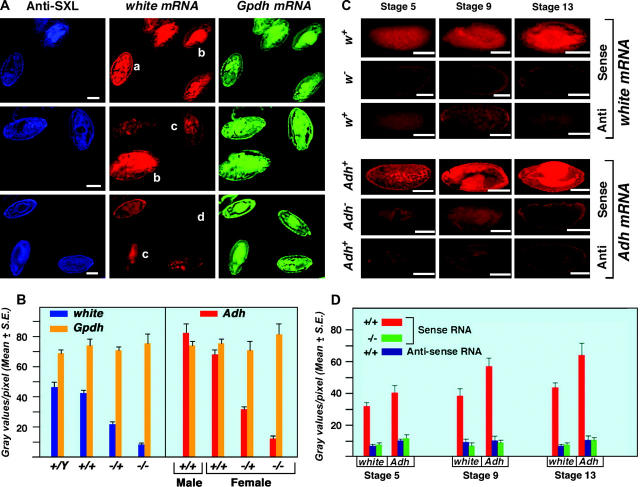
Figure 2.—
Development of a quantitative fluorescent gene expression assay. (A) Embryos stained with SXL antibodies and probed with antisense white and α-Gpdh RNAs. The anti-SXL labeling (blue) distinguishes the sex of the embryos. Females express SXL while males do not. The w mRNA hybridization was visualized using Cy3-conjugated UTP incorporated into the antisense probe. α-Gpdh mRNA was hybridized with an antisense probe labeled with Cy5-conjugated UTP. The pattern of Canton-S males and females is shown at the top. A segregating progeny of normal males and w67c23/+ heterozygous females, generated by crossing Canton-S females by males of a white deficiency strain (w67c23), are in the middle. A population of w−/+ female and w−/Y male embryos, which result from the reciprocal cross, are shown at the bottom. The genotype of each embryo is (a) Canton-S female, (b) Canton-S male, (c) w−/+ female, and (d) w− male. Bars, 50 μm. (B) A quantitative measurement of w, Adh, and α-Gpdh mRNA in embryos segregating for normal or null alleles of white or Adh, respectively. The white genotypes are described above. The heterozygous Adh normal/null genotype was produced by crossing Adhfn6 by Canton-S wild type, whereas the null homozygotes were obtained from an Adhfn6/ Adhfn6 stock. Normal males and females were obtained from the Canton-S stock. The Adh mRNA was detected with a digoxygenin-labeled antisense probe followed by incubation with Cy5-conjugated anti-dioxygenin antibodies. The same specific probe preparation for each of the three genes analyzed was used on all genotypes. Twenty-one embryos at approximately stage 13 were analyzed for each genotype. The amount of white, Adh, and α-Gpdh mRNA in each respective embryo was estimated by measuring the gray values per pixel using Metamorph software. The results indicate that the amount of mRNA detected by this technique is proportional to functional allele dosage and that a twofold difference in expression can be robustly distinguished. The results also illustrate that the quantitation of different genes (w vs. α-Gpdh) is independent of each other. (C) Comparison of null and antisense determinations of background. Normal embryos were probed for sense and antisense RNAs of white and Adh. In parallel, embryos homozygous for the respective null alleles were probed for mRNA. Embryos from three developmental stages are shown. The results illustrate that probing for antisense RNA is an effective background measurement. (D) Histograms of the comparison of background determinations via antisense probing or a null allele. The same digoxygenin-labeled probe preparation was used for analysis of normal and null genotypes for the respective genes. Twenty-one embryos were used to determine each mean. The results illustrate that the background determination by the two methods is comparable at the three developmental stages examined.