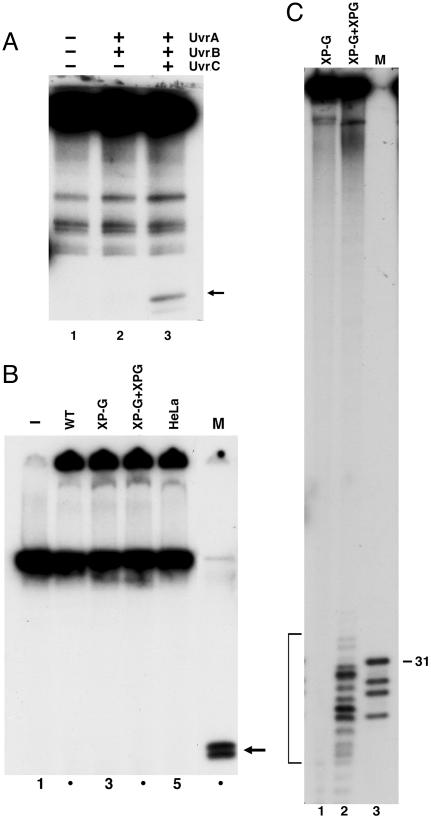
Fig. 2.
Excision of DNA–protein crosslinks by E. coli and human excision nucleases. (A) Duplex (140 bp) containing T4-pdg crosslinked to a unique AP site (0.6 nM) was incubated with 50 nM UvrA, 200 nM UvrB, and 200 nM UvrC for 60 min at 30°C, as indicated. SDS/PAGE loading buffer was added to reaction mixtures that were heated 5 min at 37°C before resolution by 12% SDS/PAGE. The closed arrow to the right identifies the band corresponding to excision product. (B) pIBI24 containing T4-pdg crosslinked to a unique AP site was incubated without (lane 1) or with 50–60 μg of extracts prepared from wild-type CHO cells (WT, AA8), XP-G mutant CHO cells (UV135), mutant extract complemented with XPG protein (XP-G + XPG), and HeLa cells for 60 min at 30°C (lanes 2–5). SDS/PAGE loading buffer was added to reaction mixtures that were heated 5 min at 95°C before resolution by 12% SDS/PAGE. Marker (M) DNA was generated by BamHI + XhoI digestion of the substrate: T4-pdg crosslinked to 29 mer is approximately the size of the predicted excision product. (C) Effect of human excision nuclease on amino acid–DNA crosslink. pIBI24–T4-pdg DNA was treated with proteinase K to generate a plasmid containing a crosslinked amino acid (pIBI24–AA). DNA (0.3 nM) was incubated with XP-G mutant CHO cell extract (UV135) or CHO-UV135 extract complemented with recombinant XPG for 60 min at 30°C. Bracket indicates excision products (13.5% of the substrate). Lane 3 (M) is pIBI24–AA digested with BamHI + XhoI to release a 29 mer containing the damage; this species migrates as a 31 mer, and faster-migrating species are reduced AP site 29 mer and fragments resulting from digestion of incomplete ligation products.