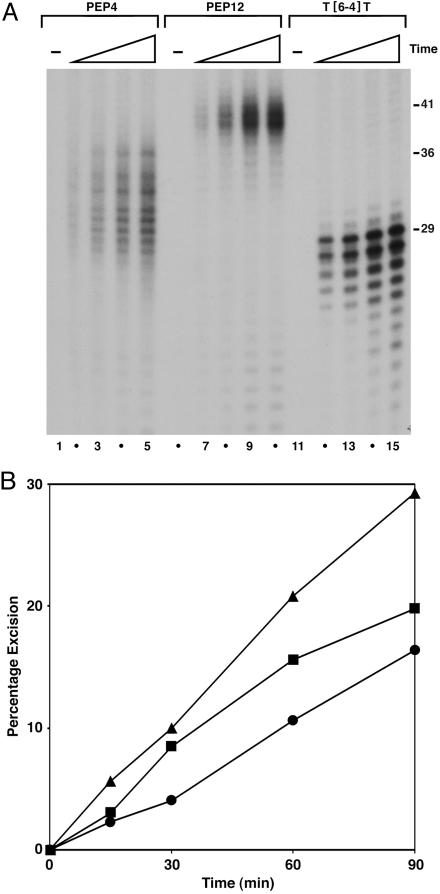
Fig. 5.
Kinetics of excision by CHO cell-free extracts. Substrates (0.5 nM) were incubated at 30°C with 50 μg of XP-G (UV135) cell extract complemented with 15 ng of recombinant XPG. Aliquots were removed at 15-, 30-, 60-, and 90-min time points, deproteinized, and precipitated with ethanol. Recovered DNA was resolved in 10% sequencing gel, visualized by autoradiography, and quantified by PhosphorImager analysis. (A) Autoradiogram of representative experiment; only the bottom segment of the autoradiogram is shown. Substrates were DNA–KWKK crosslink (PEP4, lanes 1–5), DNA–KFHEKHHSHGRY crosslink (PEP12, lanes 6–10), and T[6-4]T photoproduct (lanes 11–15). Numbers to the right indicate size (in nucleotides) at which the largest major excision product migrated: 41 (PEP12), 36 (PEP4), and 29 (T[6-4]T). (B) Quantitative analysis of excision. Average values of excision from results shown in A and a second experiment performed under the same conditions are plotted. Triangles, T[6-4]T; squares, PEP12; circles, PEP4.