Abstract
BLM encodes a member of the highly conserved RecQ DNA helicase family, which is essential for the maintenance of genome stability. Homozygous inactivation of BLM gives rise to the cancer predisposition disorder Bloom’s syndrome. A common feature of many RecQ helicase mutants is a hyperrecombination phenotype. In Bloom’s syndrome, this phenotype manifests as an elevated frequency of sister chromatid exchanges and interhomologue recombination. We have shown previously that BLM, together with its evolutionarily conserved binding partner topoisomerase IIIα (hTOPO IIIα), can process recombination intermediates that contain double Holliday junctions into noncrossover products by a mechanism termed dissolution. Here we show that a recently identified third component of the human BLM/hTOPO IIIα complex, BLAP75/RMI1, promotes dissolution catalyzed by hTOPO IIIα. This activity of BLAP75/RMI1 is specific for dissolution catalyzed by hTOPO IIIα because it has no effect in reactions containing either Escherichia coli Top1 or Top3, both of which can also catalyze dissolution in a BLM-dependent manner. We present evidence that BLAP75/RMI1 acts by recruiting hTOPO IIIα to double Holliday junctions. Implications of the conserved ability of type IA topoisomerases to catalyze dissolution and how the evolution of factors such as BLAP75/RMI1 might confer specificity on the execution of this process are discussed.
Keywords: Bloom’s syndrome, Holliday junction dissolution, topoisomerase III, sister chromatid exchanges
The RecQ family of DNA helicases is essential for the maintenance of genome stability (1). The human genome contains five RecQ helicase genes. Mutations in three of these genes give rise to clinically defined cancer predisposition disorders (2). One of these disorders is Bloom’s syndrome (BS), which is caused by biallelic mutations in the BLM gene (3). The BLM protein is a 3′–5′ DNA helicase that processes a broad range of structurally diverse DNA substrates (4–7). These substrates include DNA structures that arise during homologous recombination, such as D-loops and Holliday junctions (5, 6). These structures are of particular relevance to the BS phenotype because BS cells display elevated levels of homologous recombination (8). This hyperrecombination phenotype is also a feature of Saccharomyces cerevisiae and Schizosaccharomyces pombe mutants defective in their respective BLM orthologs, SGS1 and rqh1+ (9–11). In the case of BS cells, recombination events are particularly apparent between sister chromatids, and such recombination events are termed sister chromatid exchanges (SCEs) (8). These exchanges arise primarily as a consequence of crossing-over during the processing of recombination intermediates (12).
BLM exists in a complex with topoisomerase IIIα (hTOPO IIIα), a type IA topoisomerase (13, 14). This complex is evolutionarily conserved, and functional and/or physical interactions between RecQ helicases and type IA topoisomerases have also been demonstrated in bacteria and yeast (9, 15–17). Two type IA topoisomerases are found in Escherichia coli (Top1 and Top3) and in mammals (TOPO IIIα and TOPO IIIβ), whereas budding yeast contains a single type IA topoisomerase (Top3) (18). In unicellular organisms, mutations in type IA topoisomerases give rise to a hyperrecombination phenotype, indicating that RecQ helicase/type IA topoisomerase complexes act during homologous recombination (19, 20). Indeed, many aspects of the phenotypes of RecQ helicase and type IA topoisomerase mutants can be suppressed by mutations in the RAD52 epistasis group of recombinational repair genes, suggesting a role for these two classes of proteins in the processing of recombination intermediates (11, 21, 22). In budding yeast, Sgs1 and its binding partner Top3 have been shown to suppress the formation of crossovers during homologous recombination-mediated repair of a DNA double-strand break, a process that would suppress SCEs (23). Consistent with this finding, we have shown that BLM and hTOPO IIIα cooperate to convert double Holliday junction (DHJ) structures exclusively into noncrossover products by a mechanism termed dissolution (24).
Recently, BLAP75 was identified as a third, evolutionarily conserved, component of the BLM/hTOPO IIIα complex (25, 26). In yeast, the ortholog of BLAP75, Rmi1, was identified independently by two groups as a subunit of the Sgs1–Top3 complex (27, 28). In human and yeast cells, BLAP75 and Rmi1, respectively, are required for the stability of the RecQ helicase/type IA topoisomerase complex in which each protein resides (26–28). BLAP75 and Rmi1 contain a conserved nucleic acid recognition motif, termed the oligonucleotide/oligosaccharide-binding (OB)-fold domain, and are, therefore, predicted to bind DNA (26–28). Indeed, recent data indicate that Rmi1 is a DNA-binding protein with a preference for Holliday junctions (27). However, the precise roles of BLAP75 and Rmi1 are unknown, although genetic analyses indicate that their function is closely associated with the activity of the RecQ helicase/Topo III complex. For example, RNA interference-mediated down-regulation of BLAP75 results in an increase in the frequency of SCEs (26). In yeast, rmi1Δ mutants phenocopy top3Δ cells and display an array of genetic interactions similar to that seen with TOP3 (27, 28). In particular, mutations in SGS1 can suppress many of the phenotypes of both rmi1 and top3 mutants, suggesting that Rmi1 acts downstream of Sgs1 alongside Top3 in the resolution of recombination intermediates (27, 28). To avoid confusion over nomenclature, we will henceforth refer to the human ortholog of Rmi1 as hRMI1 instead of BLAP75.
In this study, we have identified a previously unrecognized biochemical function of hRMI1 by showing that hRMI1 cooperates with BLM and hTOPO IIIα to catalyze DHJ dissolution. Moreover, we show that this role of hRMI1 is mediated through a specific interaction with hTOPO IIIα and that hRMI1 promotes the recruitment of hTOPO IIIα to DHJs.
Results and Discussion
hRMI1 Is Required for Efficient DHJ Dissolution.
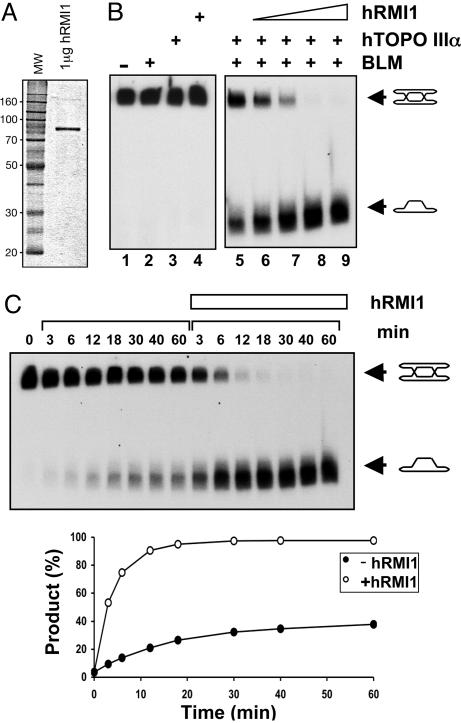
We proposed previously that DHJ dissolution represents a mechanism that eliminates crossing-over during recombination and hence suppresses the formation of SCEs (24). The physical association of hRMI1 with BLM, together with the elevated SCE frequency seen after RNA interference-mediated suppression of hRMI1 expression (26), prompted us to examine whether hRMI1 might have an influence on BLM/hTOPO IIIα-mediated dissolution. Recombinant hRMI1 was expressed and purified to near homogeneity from E. coli cells as described in Methods (Fig. 1A). The dissolution reaction was performed by using a previously described substrate, termed DHJ, that consists of two interlinked circular oligonucleotides that, when annealed, form a DHJ structure (24). Dissolution of DHJ results in the release of two intact circular oligonucleotides (24). As shown previously, the DHJ structure is subject to dissolution by the combined action of BLM and hTOPO IIIα (Fig. 1B, lane 5). Interestingly, although hRMI1 alone had no effect on DHJ, we found that the addition of hRMI1 stimulated, in a concentration-dependent manner, BLM and hTOPO IIIα-mediated dissolution (Fig. 1B). Time-course experiments indicated a >10-fold increase in the rate of dissolution when hRMI1 was included in the reaction (Fig. 1C). This stimulatory effect of hRMI1 was seen with recombinant protein purified from either E. coli (Fig. 1) or insect cells (data not shown), thereby eliminating the possibility that the stimulatory effect of the hRMI1 preparation was due to the presence of a contaminating protein derived from the host cell. hRMI1 incubated with either BLM alone or hTOPO IIIα alone generated no dissolution product (Fig. 2A, lanes 9 and 14 and Fig. 2B, lanes 9 and 14), indicating that hRMI1 causes a bona fide stimulation of the dissolution reaction as opposed to being able to functionally substitute for either BLM or hTOPO IIIα.
Fig. 1.
hRMI1 stimulates BLM/hTOPO IIIα-mediated dissolution. (A) Coomassie blue-stained polyacrylamide gel showing recombinant hRMI1 purified from E. coli cells. (B) Dissolution reactions containing BLM (2.5 nM, lanes 2 and 5–9), hTOPO IIIα (83 nM, lanes 3 and 5–9), and a 2-fold dilution series of hRMI1 (83 nM, lane 6; 166 nM, lane 7; 332 nM, lane 8; and 664 nM, lanes 4 and 9) as indicated above the lanes. The positions of DHJ and the detectable dissolution product are indicated on the right. (C Upper) Time course of dissolution reactions containing BLM (2.5 nM) and hTOPO IIIα (83 nM) with or without hRMI1 (664 nM) as indicated by the white bar above the lanes. The positions of DHJ and the detectable dissolution product are indicated on the right. (C Lower) Quantification of the dissolution product as a function of time in the presence or absence of hRMI1 as indicated.
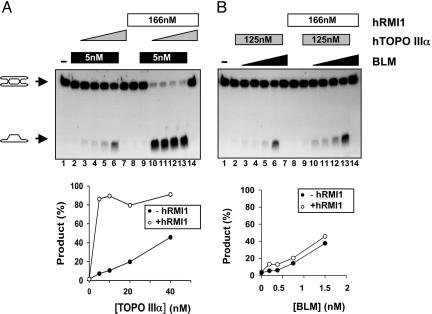
Fig. 2.
hRMI1 specifically stimulates the hTOPO IIIα component of dissolution reactions. (A Upper) Dissolution reactions containing fixed concentrations of BLM and hRMI1 as indicated by the black and white bars, respectively, and a 2-fold dilution series of hTOPO IIIα (highest concentration of 40 nM, indicated by the gray triangles). The positions of DHJ and the detectable dissolution product are indicated on the left. (A Lower) Quantification of the dissolution product in lanes 2–6 (−hRMI1) and lanes 9–13 (+hRMI1) as a function of hTOPO IIIα concentration. (B Upper) Dissolution reactions containing fixed concentrations of hTOPO IIIα and hRMI1, as indicated by the gray and white bars, respectively, and a 2-fold dilution series of BLM (highest concentration of 1.5 nM, indicated by black triangles). (B Lower) Quantification of the dissolution product in lanes 2–6 (−hRMI1) and lanes 9–13 (+hRMI1) as a function of BLM concentration.
hRMI1 Specifically Stimulates hTOPO IIIα in the Catalysis of DHJ Dissolution.
Dissolution is absolutely dependent on both BLM and hTOPO IIIα (24). A number of possibilities existed, therefore, for how hRMI1 might act to stimulate dissolution: (i) hRMI1 specifically stimulates BLM; (ii) hRMI1 specifically stimulates hTOPO IIIα; or (iii) hRMI1 stimulates the concerted action of BLM and hTOPO IIIα on DHJ structures. To distinguish between these possibilities, we performed dissolution reactions under conditions in which either BLM or hTOPO IIIα was present at limiting concentrations to restrict the extent of dissolution. When hRMI1 was added to reactions in which the concentration of hTOPO IIIα was limiting, a strong stimulatory effect could be observed (Fig. 2A). At lower concentrations of hTOPO IIIα, the extent of dissolution was >10-fold greater in the presence of hRMI1 than in its absence (Fig. 2A). In contrast, hRMI1 had no significant effect in reactions in which the concentration of BLM limited the extent of dissolution (Fig. 2B). Thus, hRMI1 specifically stimulates the hTOPO IIIα component of the dissolution reaction. Two observations are consistent with this result. First, in yeast, mutation of RMI1 generates a phenocopy of a top3 mutation that can be suppressed by deletion of SGS1 (27, 28). This observation suggests that Rmi1 acts downstream of Sgs1 in conjunction with Top3. Second, dissolution catalyzed by BLM and hTOPO IIIα does not require an evolutionarily conserved hTOPO IIIα interaction domain located in the N-terminal domain of BLM (29). This observation suggests that BLM and hTOPO IIIα might act sequentially to catalyze dissolution. Under these circumstances, it is possible, therefore, that hRMI1 exerts its effects directly on hTOPO IIIα.
hRMI1 Cannot Stimulate Dissolution Catalyzed by Other Type IA Topoisomerases.
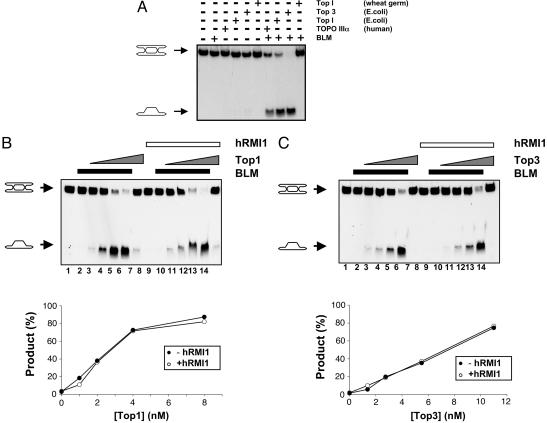
Next, we addressed the mechanism by which hRMI1 promotes dissolution. Because hRMI1 acts on the hTOPO IIIα component of dissolution, we surmised that hRMI1 might mediate its effect either through some modulation of the DNA substrate that facilitates hTOPO IIIα-mediated strand passage activity or through a direct stimulation of hTOPO IIIα activity. To address the former possibility, we first determined whether other type IA topoisomerases could catalyze dissolution in conjunction with BLM. The process of dissolution has a specific requirement for BLM, because neither E. coli RecQ nor three other human RecQ helicases (WRN, RECQ1, and RECQ5β) could substitute for BLM in dissolution reactions (29). However, in contrast to BLM, we found that hTOPO IIIα could be replaced by either of the E. coli type IA topoisomerases, Top1 and Top3 (Fig. 3A). As shown in ref. 29, wheat germ topoisomerase 1, which is a type IB topoisomerase, could not substitute for hTOPO IIIα, indicating that dissolution displays a specific requirement for type IA topoisomerase activity (Fig. 3A). Importantly, dissolution catalyzed by E. coli Top1 and Top3 was BLM-dependent, indicating that the reaction catalyzed by each of the bacterial topoisomerases is mechanistically similar to dissolution carried out by hTOPO IIIα. The ability of E. coli Top1 to substitute for hTOPO IIIα is consistent with the observation that overexpression of this protein in yeast can suppress some of the phenotypes of top3 mutant cells (20).
Fig. 3.
hTOPO IIIα can be replaced by bacterial type IA topoisomerases in dissolution reactions. (A) Dissolution reactions containing BLM (5 nM) and hTOPO IIIα (250 nM), E. coli Top1 (50 nM), E. coli Top3 (15 nM), or wheat germ topoisomerase I (10 units), as indicated above the lanes. The positions of DHJ and the detectable dissolution product are indicated on the left. (B Upper) Dissolution reactions containing BLM (5 nM), as indicated by the black bars, hRMI1 (166 nM), as indicated by the white bar, and a 2-fold dilution series of E. coli Top1 (highest concentration of 8 nM), as indicated by the gray triangles. The positions of DHJ and the detectable dissolution product are indicated on the left. (B Lower) Quantification of the dissolution product in lanes 2–6 (−hRMI1) and lanes 9–13 (+hRMI1) as a function of E. coli Top1 concentration. (C Upper) Dissolution reactions containing BLM (5 nM), as indicated by the black bars, hRMI1 (166 nM), as indicated by the white bar, and a 2-fold dilution series of E. coli Top3 (highest concentration of 11 nM), as indicated by the gray triangles. The positions of DHJ and the detectable dissolution product are indicated on the left. (C Lower) Quantification of the dissolution product in lanes 2–6 (−hRMI1) and lanes 9–13 (+hRMI1) as a function of Top3 concentration.
The finding that E. coli Top1 and Top3 can catalyze dissolution in a BLM-dependent manner allowed us to test whether hRMI1 could stimulate dissolution catalyzed by type IA topoisomerases other than hTOPO IIIα. In contrast to reactions containing hTOPO IIIα, the addition of hRMI1 to reactions containing either of the bacterial type IA topoisomerases had no effect on the efficiency of dissolution (Fig. 3 B and C). We conclude, therefore, that the specific nature of the functional interaction between hRMI1 and hTOPO IIIα versus other type IA topoisomerases makes it very unlikely that the stimulatory effect of hRMI1 is mediated through modulation of the DHJ molecule itself to facilitate the strand passage activity of type IA topoisomerases. Moreover, because all of these reactions contained BLM, this observation also lends further support to the notion that hRMI1 does not mediate its stimulatory effect on dissolution through modulation of the activity of BLM.
hRMI1 Promotes hTOPO IIIα Binding to DHJs.
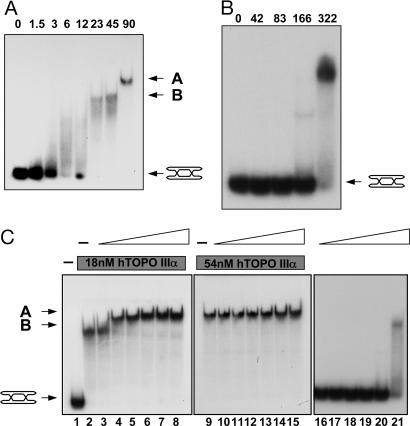
Given that the mechanism by which hRMI1 stimulates dissolution is mediated through neither a direct effect on BLM nor a modulation of the DHJ DNA to facilitate the strand passage activity of type IA topoisomerases, we investigated whether hRMI1 modulated the activity of hTOPO IIIα alone on the DHJ substrate. As stated above, hRMI1 was not able to circumvent the requirement for BLM in dissolution reactions and hence was not able to stimulate the strand passage activity of hTOPO IIIα (in the absence of BLM) on the DHJ substrate. We analyzed, therefore, whether hRMI1 mediates its stimulatory effect on dissolution by influencing the ability of hTOPO IIIα to bind to the DHJ molecule. To analyze this, we used EMSAs to assess the formation of protein–DNA complexes. hTOPO IIIα alone was found to bind the DHJ and generated multiple protein–DNA complexes whose electrophoretic mobility was inversely proportional to protein concentration. At high protein concentrations, hTOPO IIIα produced two discernible protein–DNA species, termed complexes A and B (Fig. 4A). At the highest concentration of hTOPO IIIα tested, the formation of the slowest migrating species, complex A, was predominant, whereas at lower concentrations of hTOPO IIIα, complex B was predominant (Fig. 4A). At concentrations below those required for the formation of complex B, hTOPO IIIα–DNA complexes migrated as a smear that had a reduced electrophoretic mobility compared with the unbound substrate. This profile of protein–DNA complex formation suggests that multiple molecules of hTOPO IIIα may load onto a single DHJ molecule, in a stepwise manner, giving rise to progressively lower mobility species as the hTOPO IIIα protein concentration is increased. The inability to detect discrete protein–DNA complexes at lower hTOPO IIIα concentrations suggests that at these concentrations of hTOPO IIIα, protein–DNA complexes are somewhat unstable and may dissociate during gel electrophoresis. hRMI1 alone did not bind DHJ at concentrations equivalent to that at which hTOPO IIIα caused complete retardation of the substrate. However, at higher concentrations (>166 nM) hRMI1 alone bound DHJ, generating one minor and one major protein–DNA complex (Fig. 4B). This finding is consistent with the ability of yeast RMI1 to bind single Holliday junctions and ssDNA because elements of both structures are likely to be present in the DHJ molecule.
Fig. 4.
hRMI1 promotes hTOPO IIIα binding to DHJ. (A) EMSAs using DHJ and various concentrations of hTOPO IIIα as indicated. Positions of protein–DNA complexes, designated A and B, and the unbound DHJ substrate are indicated on the right. (B) EMSAs using DHJ and various concentrations of hRMI1 as indicated. The position of the unbound DHJ substrate is indicated on the right. (C) EMSAs using DHJ, two fixed concentrations of hTOPO IIIα, as indicated by the gray bars (lanes 2–8 and 9–15), and a 3-fold dilution series of hRMI1 (highest concentration of 256 nM), as indicated by the white triangles (lanes 3–8, 10–15, and 16–21). Positions of protein–DNA complexes, designated A and B, and the unbound DHJ substrate are indicated on the left.
Next, we analyzed whether hRMI1 could influence the DNA-binding properties of hTOPO IIIα. To do this, we used two concentrations of hTOPO IIIα at which either complex A or complex B could be detected and analyzed the effect of addition of hRMI1. Interestingly, addition of hRMI1 at concentrations 80-fold below that required to show DHJ binding (3 nM) caused a decrease in the electrophoretic mobility of complex B (Fig. 4C). Moreover complex B was converted, in the presence of hRMI1, to a species that had an electrophoretic mobility that was indistinguishable from that of complex A. In contrast, addition of hRMI1 had no effect on complex A (Fig. 4C). These data suggest that hRMI1 can stimulate the stepwise loading of hTOPO IIIα onto DHJ to form complex A. Complex A likely represents a protein–DNA complex that contains the maximal number of stably bound hTOPO IIIα molecules and is therefore resistant to the stimulatory effects of hRMI1. When both proteins were incubated together with DHJ at concentrations at which either protein alone bound DHJ, no new unique complexes, dependent on the presence of both hRMI1 and hTOPO IIIα, could be detected. Indeed, at high concentrations of hRMI1, the formation of hRMI1–DHJ complexes was inhibited in the presence of hTOPO IIIα, suggesting that hRMII and hTOPO IIIα cannot simultaneously bind DHJ (Fig. 4C, compare lanes 8, 15, and 21). This finding suggests that the ability of hRMI1 to promote the loading of hTOPO IIIα onto DHJ does not require its stable association with the hTOPO IIIα–DHJ complex. Consistent with this notion is the fact that a less than 1:1 molar ratio of hRMI1:hTOPO IIIα is required for hRMI1 to load hTOPO IIIα onto DHJ (Fig. 4C).
hTOPO IIIα and hRMI1 Physically Interact.
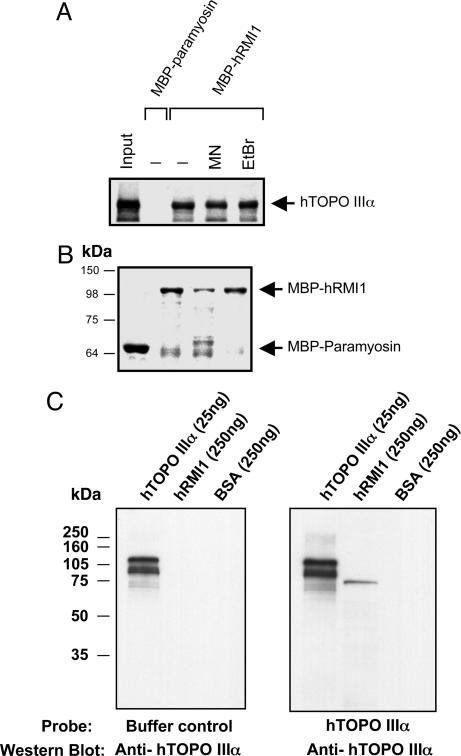
The ability of hRMI1 to load hTOPO IIIα onto DHJ without stably associating with the hTOPO IIIα–DHJ complex and at concentrations at which hRMI1 alone did not stably bind DHJ suggested that hRMI1 might exert its effect through a protein–protein interaction with hTOPO IIIα. Such an interaction might induce a conformational change in hTOPO IIIα that facilitates its loading onto DHJ structures. We therefore tested, using two independent means, whether hTOPO IIIα and hRMI1 could physically interact. First, we produced recombinant hRMI1 protein in the form of a maltose-binding protein (MBP) fusion and examined its binding to in vitro-translated hTOPO IIIα in a pull-down assay (Fig. 5A and B). hTOPO IIIα was efficiently retained by MBP-hRMI1 immobilized on amylose beads but was not efficiently retained by the MBP-paramyosin control, suggesting a physical interaction between hRMI1 and hTOPO IIIα. Inclusion of either micrococcal nuclease or ethidium bromide had little or no effect on the ability of hRMI1 and hTOPO IIIα to interact, suggesting that the interaction was not mediated by DNA (Fig. 5 A and B). Second, to confirm that the interaction between hTOPO IIIα and hRMI1 was direct and not mediated by proteins present in the in vitro translation reaction, purified hTOPO IIIα and hRMI1 were subjected to Far-Western analysis in which hRMI1 immobilized on a membrane was used to capture hTOPO IIIα (Fig. 5C). By using this methodology, hTOPO IIIα was found to specifically interact with hRMI1. We thus conclude that hRMI1 and hTOPO IIIα can directly interact independently of DNA.
Fig. 5.
Direct interaction between hRMI1 and hTOPO IIIα. (A) Binding of in vitro-translated, 35S-labeled hTOPO IIIα to MBP-paramyosin or MBP-hRMI1. Reactions were performed in the absence or presence of micrococcal nuclease (MN, 2 units) or ethidium bromide (EtBr, 100 μg/ml) as indicated. The lane labeled “input” contains 20% of the amount of total labeled hTOPO IIIα that was added to each of the other reactions. (B) Coomassie blue staining showing the amount of the indicated MBP fusion proteins present in the binding reactions shown in A. (C) Direct binding of hTOPO IIIα and hRMI1. Identical membranes containing hTOPO IIIα (positive control), hRMI1, or BSA (negative control) were incubated with either hTOPO IIIα or control buffer, as indicated below, and Western blotting for hTOPO IIIα was performed.
Concluding Remarks.
The ability of hRMI1 to stimulate DHJ dissolution is a previously unrecognized biochemical activity ascribed to this recently identified, evolutionarily conserved component of the BLM/hTOPO IIIα complex. The elevated frequency of SCEs seen in BS cells and after RNA interference-mediated suppression of hRMI1 strongly supports the notion that dissolution acts to suppress the formation of crossovers that can arise during homologous recombination (8, 26). Several lines of evidence suggest that catalysis of dissolution is highly specific for and requires the coordinate action of the components of this complex. First, there is an absolute requirement for BLM in dissolution because no other helicase tested, including several other RecQ helicases, can support the reaction (29). This requirement is, in part, mediated through the helicase and ribonuclease D C-terminal domain of BLM, which specifically acts to promote BLM binding to DHJ structures (29). Second, there is a requirement for the strand passage activity of hTOPO IIIα. In contrast to BLM, the requirement for hTOPO IIIα, at least in vitro, is less specific because other type IA topoisomerases can catalyze dissolution in conjunction with BLM. This finding is consistent with the observation that in vitro, the conserved hTOPO IIIα interaction domain located in the N-terminal domain of BLM is dispensable for dissolution. However, in vivo, there likely exists a highly specific requirement for hTOPO IIIα in dissolution, which may be conferred by at least two features: the correct subcellular localization of hTOPO IIIα (14), which does require a physical interaction of BLM and hTOPO IIIα and, as demonstrated in this study, the ability of hRMI1 to stimulate the recruitment of hTOPO IIIα to DHJ structures. It remains to be determined how hRMI1 promotes the binding of hTOPO IIIα to DHJ structures. Our data are consistent with two models, which are not necessarily mutually exclusive: (i) hRMI1 binds DHJ structures and, through a physical association, recruits hTOPO IIIα before dissociating from the DNA, and (ii) hRMI1 induces a conformational change in hTOPO IIIα, which facilitates the binding of hTOPO IIIα to DHJ structures. The generation of mutations in hRMI1 that disrupt the ability of hRMI to interact with either hTOPO IIIα or DNA will be important in distinguishing between such models. It also remains to be determined what structural features of the DHJ molecule are recognized by hRMI1 and hTOPO IIIα and how modification of the DHJ substrate by BLM might affect the functional relationship between hRMI1 and hTOPO IIIα in binding to the DHJ. Both yeast Rmi1 and hTOPO IIIα preferentially bind single-stranded DNA over double-stranded DNA. Yeast Rmi1 can also bind single Holliday junctions. To what degree these binding activities contribute to the recognition of DHJ by hRMI1 and hTOPO IIIα is currently unknown. It is possible that the full extent of the stimulatory effect of hRMI1 on hTOPO IIIα binding to DHJ requires conversion of the DHJ molecule into a yet to be identified intermediate by BLM.
The specificity of the BLM/hTOPO IIIα/hRMI1 complex to catalyze dissolution likely evolved to preclude aberrant processing of DHJs by other DNA-metabolizing enzymes, which could result in genome destabilization. Further analysis of this heteromeric DHJ processing complex will be important in furthering our understanding of the mechanism by which cells effect efficient repair of DNA double-strand breaks and damaged replication forks, both of which can give rise to DHJs (23, 24, 30–32).
Methods
Proteins.
hRMI1 was expressed and purified from E. coli cells. The hRMI1 ORF was PCR amplified from the cDNA clone AK022950 and cloned into pET15b. The resulting plasmid, hRMI1-pET15b, was transformed into BL21(λDE3). One liter of bacteria was grown at 37°C to midlogarithmic phase, and isopropyl β-d-thiogalactoside was added to give a final concentration of 0.4 mM to induce expression of hRMI1. Growth was continued overnight at 16°C. Cells were harvested and resuspended in 5 ml of 2× lysis buffer [100 mM Hepes-KOH, pH 7.5/20% (vol/vol) glycerol/20 mM sodium pyrophosphate/100 mM NaF/2 mM Na3VO4] with 2 mM PMSF and 80 μg/ml leupeptin. Water was added to a final volume of 10 ml. The cell suspension was sonicated with five pulses of 30 s with 1 min of cooling on ice between pulses. Nonidet P-40 and NaCl were added to final concentrations of 0.1% and 250 mM, respectively, and the lysate was incubated on ice for 15 min. The lysate was centrifuged for 30 min at 20,000 rpm at 4°C in a Sorvall SS34 rotor. hRMI1 was found in the insoluble fraction. Two grams of the insoluble fraction was resuspended in 40 ml of extraction/wash (E/W) buffer (50 mM sodium phosphate/6 M guanidine·HCl/300 mM NaCl, pH 7.0) and agitated until the suspension became translucent. After centrifugation at 15,000 rpm for 20 min at 4°C in a Sorvall SS34 rotor, the supernatant was added to 5 ml of TALON resin (Clontech) and mixed for 1.5 h at 4°C. The supernatant was removed, and the resin was washed three times with 50 ml of E/W buffer. The resin was transferred to a column and washed twice with 50 ml of E/W buffer, and bound proteins were eluted with 45 mM sodium phosphate/5.4 M guanidine·HCl/270 mM NaCl/150 mM imidazole (pH 7.0). The purified RMI1 was renatured by dialysis against two changes of 2.5 liters of PBS overnight at 4°C. BLM was expressed and purified as described in ref. 33. hTOPO IIIα was a gift from J.-F. Riou and H. Goulaouic (both of Aventis Pharma, France). E. coli Top1 and Top3 were gifts from K. Marians (Memorial Sloan–Kettering Cancer Center, New York). Wheat germ topoisomerase I was purchased from Promega.
Dissolution Assays.
The DHJ substrate was prepared and purified as described in refs. 24 and 34 and was added at 30 fM with the indicated proteins in reaction buffer containing 50 mM Tris·HCl (pH 7.5), 50 mM NaCl, 4 mM MgCl2, 5 mM ATP, 1 mM DTT, and 0.1 mg/ml BSA at 37°C for 60 min. Reactions were stopped by the addition of 1% SDS and 50 mM EDTA. Samples were deproteinized with proteinase K and extracted once with phenol/chloroform before being subjected to denaturing 8% PAGE. Gels were dried and subjected to phosphorimaging analysis by using a STORM 840 scanner and imagequant software (Amersham Biosciences).
DNA-Binding EMSA.
DHJ was incubated with various concentrations of protein, as indicated in the figure legends, in reaction buffer containing 20 mM triethanolamine (pH 7.5), 4 mM MgCl2, 10 μg/ml BSA, and 1 mM DTT at 37°C for 7.5 min. Samples were then subjected to 5% PAGE and visualized by autoradiography.
Pull-Down Assay.
[35S]Met-labeled hTOPO IIIα was synthesized in vitro from pcDNA3.1-hTop3 (T7) by using a TnT Quick Coupled Transcription/Translation System (Promega). MBP fusion proteins were expressed in the E. coli strain PR745. For the pull-down assays, amylose beads (NEB, Beverly, MA) were incubated with E. coli lysate containing either MBP-RMI1 or MBP-paramyosin followed by washing three times to remove unbound proteins. Upon addition of in vitro-synthesized hTOPO IIIα, with or without micrococcal nuclease or ethidium bromide, the suspensions in binding buffer [40 mM Hepes, pH 7.9/2% (vol/vol) glycerol/50 mM KCl/50 mM NaCl/5 mM MgCl2/1 mM EDTA/0.5 mM DTT/0.2 mM phenylmethanesulfonyl fluoride] were incubated at 28°C for 30 min and then incubated for another 1 h at 4°C. After four washes, bound proteins were resolved by SDS/PAGE (8%) and visualized by autoradiography or Coomassie blue staining.
Far-Western Blotting.
The indicated proteins were separated by denaturing 10% PAGE and transferred to nitrocellulose membrane. Membranes were subjected to Far-Western analysis using hTOPO IIIα as a probe and anti-hTOPO IIIα antibody as described in ref. 14.
Acknowledgments
We thank members of the Genome Integrity and Chromosome Stability groups for helpful discussions, Dr. K. Marians for E. coli Top1 and Top3, and Dr. Peter McHugh for critical reading of the manuscript. This work was funded by Cancer Research UK (I.D.H., L.W., and C.Z.B.), the Canadian Institutes of Health Research (G.W.B., J.O., and M.C.), the Intramural Research Program of the National Institute on Aging, National Institutes of Health (W.W. and J.Y.), and National Institutes of Health Extramural Grants CA91029 and CA97175 (to L.L.). G.W.B. is a research scientist of the National Institute of Canada. C.X. is supported by a fellowship from the Schissler Foundation.
Abbreviations
- BS
Bloom’s syndrome
- SCE
sister chromatid exchange
- DHJ
double Holliday junction
- MBP
maltose-binding protein
- hTOPO IIIα
human topoisomerase IIIα
- Top1 and Top3
Escherichia coli topoisomerases 1 and 3.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bachrati C. Z., Hickson I. D. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickson I. D. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 3.Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 4.Sun H., Karow J. K., Hickson I. D., Maizels N. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 5.van Brabant A. J., Ye T., Sanz M., German I. J., Ellis N. A., Holloman W. K. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 6.Karow J. K., Constantinou A., Li J. L., West S. C., Hickson I. D. Proc. Natl. Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohaghegh P., Karow J. K., Brosh R. M., Jr., Bohr V. A., Jr., Hickson I. D. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaganti R. S., Schonberg S., German J. Proc. Natl. Acad. Sci. USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart E., Chapman C. R., Al-Khodairy F., Carr A. M., Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt P. M., Hickson I. D., Borts R. H., Louis E. J. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoda E., Sasaki M. S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S. Mol. Cell. Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson F. B., Lombard D. B., Neff N. F., Mastrangelo M. A., Dewolf W., Ellis N. A., Marciniak R. A., Yin Y., Jaenisch R., Guarente L. Cancer Res. 2000;60:1162–1167. [PubMed] [Google Scholar]
- 14.Wu L., Davies S. L., North P. S., Goulaouic H., Riou J. F., Turley H., Gatter K. C., Hickson I. D. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 15.Harmon F. G., DiGate R. J., Kowalczykowski S. C. Mol. Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin A., Wang S. W., Toda T., Norbury C., Hickson I. D. Nucleic Acids Res. 1999;27:4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maftahi M., Han C. S., Langston L. D., Hope J. C., Zigouras N., Freyer G. A. Nucleic Acids Res. 1999;27:4715–4724. doi: 10.1093/nar/27.24.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L., Hickson I. D. Cell. Mol. Life Sci. 2001;58:894–901. doi: 10.1007/PL00000909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schofield M. A., Agbunag R., Michaels M. L., Miller J. H. J. Bacteriol. 1992;174:5168–5170. doi: 10.1128/jb.174.15.5168-5170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 21.Oakley T. J., Goodwin A., Chakraverty R. K., Hickson I. D. DNA Repair (Amst.) 2002;1:463–482. doi: 10.1016/s1568-7864(02)00032-0. [DOI] [PubMed] [Google Scholar]
- 22.Shor E., Gangloff S., Wagner M., Weinstein J., Price G., Rothstein R. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ira G., Malkova A., Liberi G., Foiani M., Haber J. E. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L., Hickson I. D. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 25.Meetei A. R., Sechi S., Wallisch M., Yang D., Young M. K., Joenje H., Hoatlin M. E., Wang W. Mol. Cell. Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin J., Sobeck A., Xu C., Meetei A. R., Hoatlin M., Li L., Wang W. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen J. R., Nallaseth F. S., Lan Y. Q., Slagle C. E., Brill S. J. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G. A., Boone C., Brown G. W. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu L., Lung Chan K., Ralf C., Bernstein D. A., Garcia P. L., Bohr V. A., Vindigni A., Janscak P., Keck J. L., Hickson I. D. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heyer W. D. Curr. Biol. 2004;14:R56–R58. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 31.McGlynn P., Lloyd R. G. Nat. Rev. Mol. Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 32.Saleh-Gohari N., Bryant H. E., Schultz N., Parker K. M., Cassel T. N., Helleday T. Mol. Cell. Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karow J. K., Chakraverty R. K., Hickson I. D. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 34.Fu T. J., Tse-Dinh Y. C., Seeman N. C. J. Mol. Biol. 1994;236:91–105. doi: 10.1006/jmbi.1994.1121. [DOI] [PubMed] [Google Scholar]