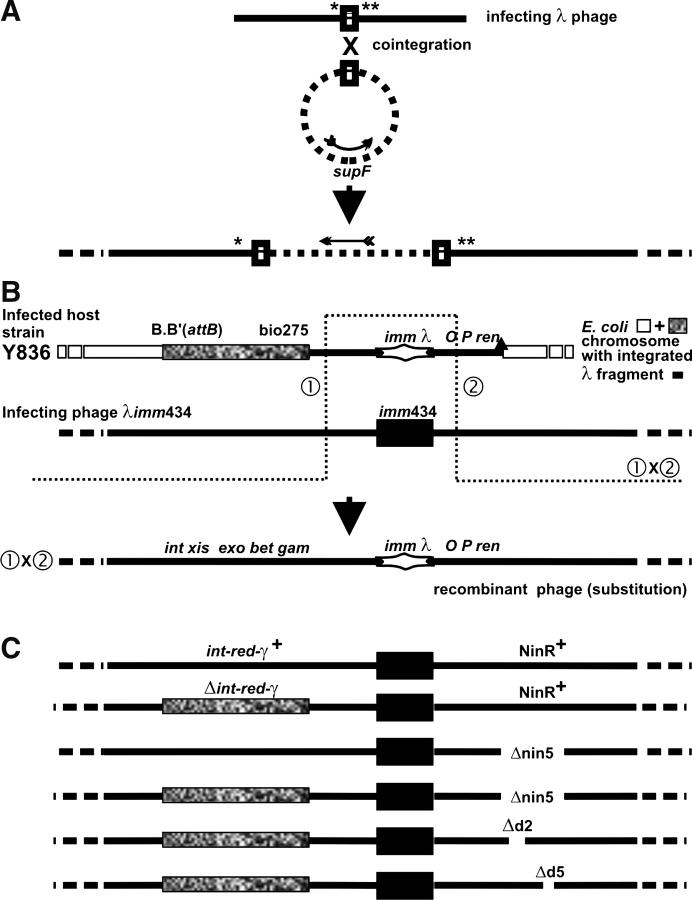
Figure 1.—
(A) Homologous recombination between common sequence (rectangle) in phage and plasmid forming a phage-plasmid co-integrate structure (see text). Shen and Huang (1986) and Hollifield et al. (1987) used infecting phages that carried amber mutations in both λ genes A and B and were deleted for recombination functions int-xis-exo-bet-gam and the genes within the NinRλ region, which were substituted by another set of genes from NinΦ80. (B) Phage-prophage marker rescue of the immλ genes from a cryptic prophage (lacking a cos site) within the bacterial chromosome by an infecting imm434 phage. The open rectangles are DNA strands of a circular E. coli chromosome; solid lines are λDNA, as infecting phage or as cryptic prophage within E. coli chromosome; the solid rectangle is imm434; the stretched diamond structure is immλ; the stippled rectangle is bio275 substitution of E. coli DNA for the λ prophage genes int-cIII in the Y836 chromosome (and in infecting phages in C); the solid triangle on map of Y836 represents the Δ431 deletion removing rightward of λ prophage genes and adjacent chromosomal genes. The actual λ bases that were deleted or substituted are described in materials and methods. The drawing is to scale, except that the sizes of the imm434 and immλ intervals were made equal when imm434 was smaller by ∼1.2 kb. (C) Maps of infecting phages used in Tables 2–5. The deletion nin5 on infecting phage encroached on the interval of λ base homology to the right of the imm434 marker (B) between the cryptic prophage and the infecting phage, reducing it from ∼2519 (or 2565) bp (with Δ431 endpoint) to ∼2257 bp. The bio275 substitution on the infecting phage increased base homology to the left of the imm434 marker: the 2281-bp λ homology was unchanged, but homology was increased by the size of the bio275 substitution (see Hayes et al. 1990) carried on both prophage and infecting Δint-red-gam phage.