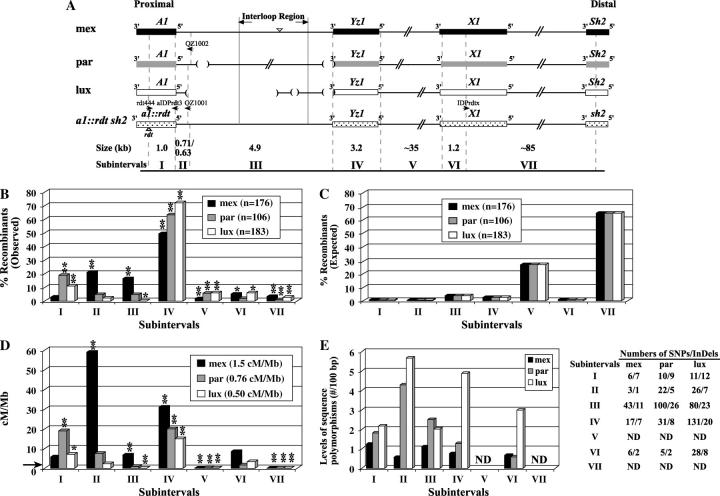
Figure 2.—
Distributions of recombination events and sequence polymorphisms across the a1-sh2 intervals of the mex, par, and lux haplotypes. (A) Comparisons of the structures of the three teosinte A1 Sh2 haplotypes relative to the maize a1::rdt sh2 haplotype. Genes are indicated as boxes. The polymorphisms shared by teosinte haplotypes relative to the a1::rdt sh2 haplotype that were used to define the subintervals are indicated by shaded dashed lines. Subintervals I, II, IV, and VI were completely sequenced for all haplotypes. Subinterval III was completely sequenced for the mex, lux, and a1::rdt sh2 haplotypes and partially sequenced for the par haplotype. Large InDeLs in subinterval III are indicated by triangles (insertions) and parentheses (deletions). The rdt transposon insertion is indicated by a triangle. Large InDeLs in other subintervals are not shown. Haplotype-specific IDP primers used to map recombination breakpoints are indicated by horizontal arrows. The sizes of each subinterval are based on those of the a1::rdt sh2 haplotype that is common among stocks carrying the mex, par, and lux haplotypes. Because no sequence polymorphisms are shared by all three haplotypes at the distal ends of subintervals II, the size of subinterval II-mex (0.71 kb) differs slightly from the sizes of subintervals II-par and II-lux (0.63 kb). Figure not to scale. (B) Observed percentages of recombinants that resolved in each subinterval. (*) and (**) indicate significant differences between the rates of recombination per megabase based on the observed recombination breakpoints mapped to subintervals and the corresponding average rates per megabase across the a1-sh2 interval of each haplotype at the 0.05 and 0.01 levels, respectively. (C) Percentages of recombinants expected to resolve in each subinterval based on a random distribution across the a1-sh2 interval. (D) Recombination rates per megabase in subintervals. The indicated average rates of recombination per megabase across the a1-sh2 interval in each of the three stocks were calculated on the basis of the physical size (∼130 kb) of the common a1::rdt sh2 haplotype carried in all stocks. The horizontal arrow indicates the average recombination rate per megabase of the maize genome (2.1 cM/Mb). (E) Levels of sequence polymorphisms (no./100 bp) between each A1 Sh2 haplotype and the a1::rdt sh2 haplotype. Numbers of SNPs/InDeLs in each subinterval are presented. Values for subinterval III-par were calculated using only the sequenced portions of this subinterval. ND, not determined.