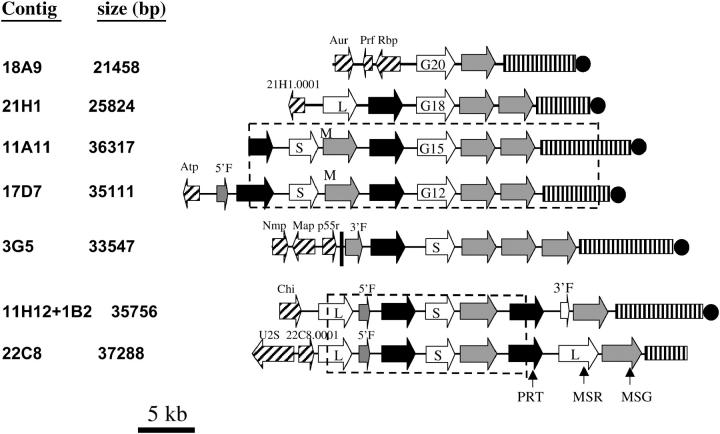
Figure 1.—
Maps of gene clusters. Arrows represent ORFs and point in the direction of transcription. Nonhatched arrows represent members of the PRT1 (solid arrows), MSR (open arrows), and MSG (shaded arrows) gene families. Rectangles with vertical lines represent subtelomeres. Solid circles represent telomeres. All features except the telomere are drawn to scale. M indicates the MSG gene that contained two point mutations. 5′F and 3′F indicate ORFs corresponding to the 5′ and 3′ ends, respectively, of either an MSG (shaded arrow) or an MSR (open arrow) gene. G12, G15, G18, and G20 indicate the MSR genes that have a poly(G) mononucleotide tract of the size denoted by the numeral. S and L indicate short and long MSR genes. The dashed-line boxes enclose regions that were at least 99% identical. The hatched arrows represent unique ORFs that had the following presumed functions, BLAST hits, and FASTA E-values: Aur1 (inositol phosphoryl-ceramide synthase, EMBL accession AF076692, 2.9 e-67), Prf (prefoldin-related, NCBI REFSEQ accession XM_331039.1, 3 e-21), Rbp (RNA binding, UniProt YAS9 SCHPO_Q10145, 7.6 e-8), 21H1.0001 (unknown function, no hits), Atp (P-type cation-pumping ATPase, NCBI accession NP_595246, 1 e-6), Nmp (nuclear migration, NCBI accession EAK84394, 2 e-61), Map (microtubule associated protein, NCBI accession XP_323888, 3 e-17), P55 (peptide similar to the p55 antigen of P. carinii, NCBI accession AAQ06671, 4 e-8), Chi (chitin synthesis, NCBI accession NP_013434, 0.084), U2S (U2 snRNP, NCBI accession NP_594538, 0), and 22C8.0001 (unknown function, NCBI accession Q09895, 6.2 e-49). All of the hatched ORFS (except Prf, Rbp, and 22C8.0001, which were not analyzed) hybridized to a single chromosome. Aur1 and Chi mapped to a 440-kb chromosome. Nmp, Map, and p55 mapped to a 290-kb chromosome. Genes 21H1.0001, Atp, and U2S mapped to chromosomes of 680, 620, and 550 kb, respectively.