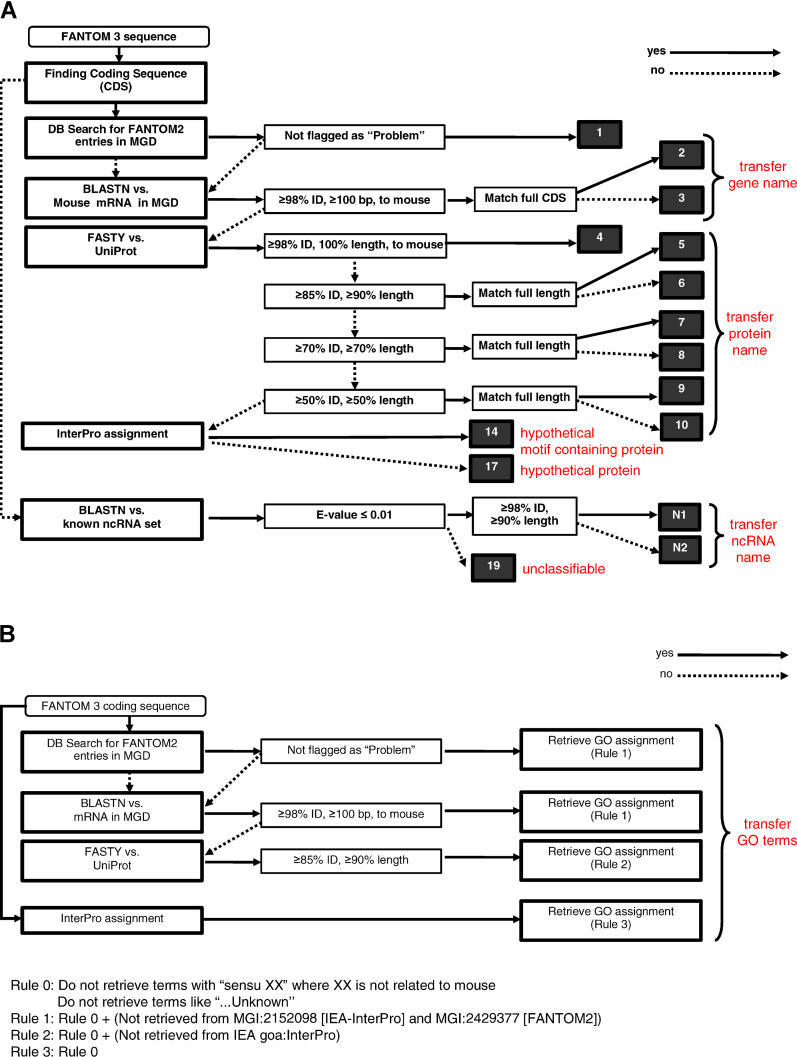
Figure 1. Annotation Pipelines for Transcript Description and for GO Terms.
(A) Pipeline for transcript description. Query sequences falling into categories (black boxes) 1–3 were assigned the description of the matched target sequence DNA entry in MGI symbols, and synonyms were also transferred to our annotation database. Queries falling into categories 4–10 were assigned a transcript description corresponding to the matched protein name. For query sequences falling into category 5 or 6, the keyword “homolog” was appended to the matching protein name. Sequences assigned to category 7 or 8 were denoted with the prefix “similar to” attached to the target sequence name. The prefix “weakly similar” was used to identify sequences assigned to category 9 or 10. For all sequences in categories 5–10, the name of the organism corresponding to the matched protein was appended to the assigned transcript description. If a query was assigned to category 14, its transcript description was “hypothetical [InterPro domain name] containing protein.” Query sequences assigned to category 17 and 19 were annotated as “hypothetical protein” and “unclassifiable,” respectively. Query sequences grouped into category N1 or N2 were assigned the description of the matched target ncRNA entry. For query sequences falling into category N2, the keyword “homolog of” was appended to the matching ncRNA name.
(B) Pipeline for GO terms.
DB, database.