Figure 1. DNA Methylation Is Not Necessary for Silencing.
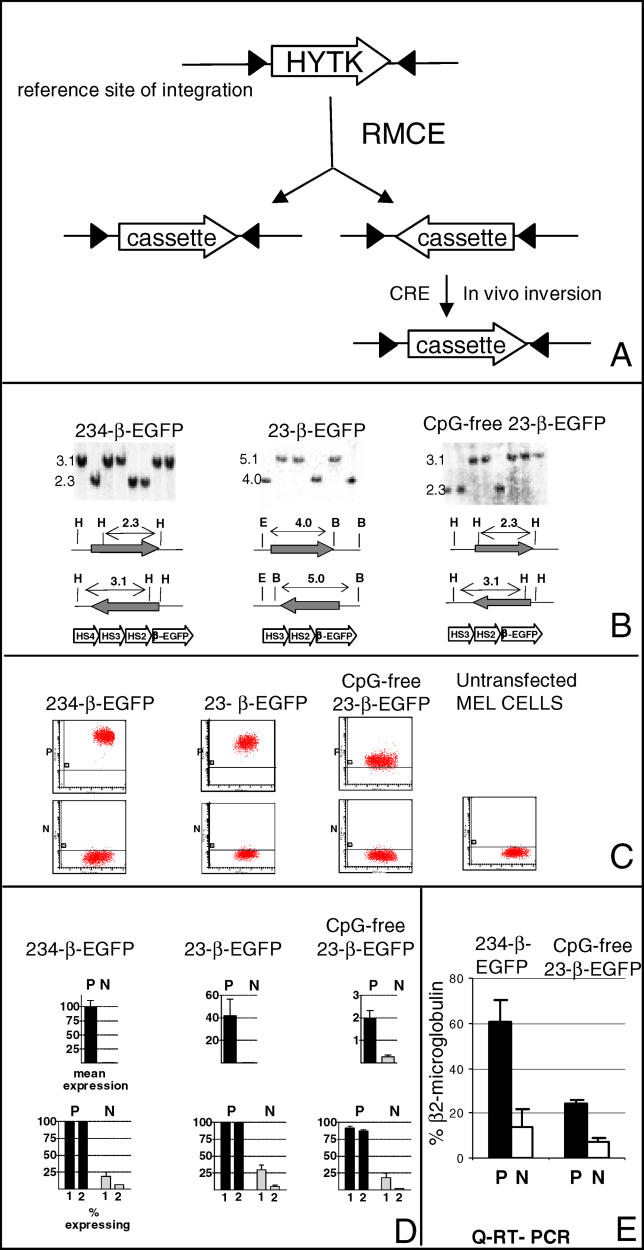
(A) Experimental system: reference integration sites are created by insertion in MEL cells of the HYTK gene flanked by two inverted L1Lox sites (black triangles). The HYTK gene can then be exchanged for any other cassettes by transfection of a Cre expression plasmid and of plasmids containing the cassette of interest. Clones with the cassettes in both orientations are obtained. Inserted cassettes can be inverted in vivo by re-expression of Cre.
(B) Southern blots demonstrating insertion of cassettes 234-β-EGFP, 23-β-EGFP, and CpG-free 23-β-EGFP. For each cassette, multiple clones in each orientation are shown.
(C) FACS analysis of typical clones in the P and the N orientations performed 6 wk post-integration. x-axis, forward-scatter; y-axis, EGFP fluorescence. All three cassettes are rapidly silenced in the N orientation, demonstrating that DNA methylation is not necessary for silencing at the RL4 locus.
(D) Quantitative analysis of expression. Top histograms: Plots of normalized mean EGFP fluorescence (± SD) of three to five clones analyzed 2-mo post-integration in the P and N orientations. EGFP fluorescence of all cassettes is normalized to expression of cassette 234-β-EGFP in the P orientation at RL4. The CpG-free cassette in the P orientation is expressed at about 5% of the efficiency of the CpG-containing 23-β-EGFP cassette and 2% of the efficiency of the 234-β-EGFP cassette. Bottom histograms, percent of cells expressing EGFP, 1- and 2-mo post-integration. The cut-off between expressing and non-expressing cells is placed using untransfected MEL cells as controls.
(E) Quantitative RT-PCR analysis of CpG-free cassette 23-β-EGFP and CpG-containing cassette 234-β-EGFP at RL4. Results are normalized to levels of β-2-micro-globulin. At the RNA level, the difference in expression between the CpG-containing and CpG-free cassettes is less than 3-fold.
N, non-permissive orientation; P, permissive orientation.