Figure 6:
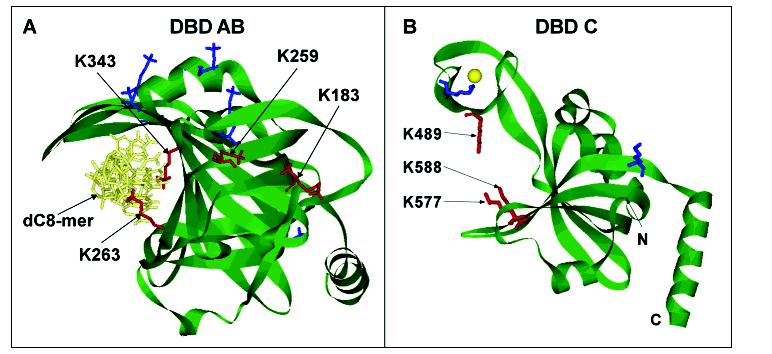
Structural exhibition of modified lysines in DBD-AB and DBD-C. (A) Structure of the DBD-AB-ssDNA complex (13). Lysine residues in the structures are presented in stick representation. K263 and K343 are found in the binding clefts of DBD-A and DBD-B, respectively, and are protected from modification by direct contact with the dC8mer. Lysine residues K183 and K259 are not in direct contact with the dC8mer but are protected from modification when ssDNA is present. (B) Structure of DBD-C (21). Lysine residues K489, K577, and K588 are located in the binding cleft of DBD-C and are protected from biotin modification when ssDNA is present, indicating direct contact with ssDNA as it transverses the binding cleft. Biotin-modified lysine residues without protection are shown in blue while the lysine residues protected from modification in the presence of ssDNA are shown in red in each structure.
