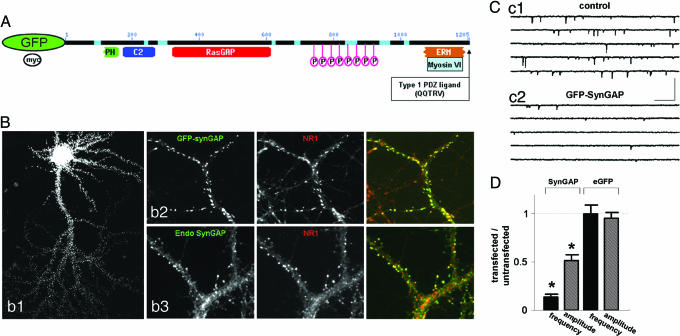
Fig. 1.
SynGAP overexpression results in decreased synaptic function. (A) Schematic of GFP-SynGAP fusion protein outlining various domains of interest. (B) Expression of GFP-SynGAP in cultured neurons. (Bb1) Low magnification of a cultured neuron expressing GFP-SynGAP for 16 h. (Bb2) GFP-SynGAP-expressing neurons were immunolabeled for GFP and NR1. (Bb3) Similar neurons were also labeled with antibodies detecting endogenous SynGAP (endo-SynGAP) and NR1. (Cc1) Whole-cell patch-clamp recording from an untransfected (control) cultured neuron. Recording solution allowed isolation of mEPSCs only from AMPARs. (Cc2) Whole-cell patch-clamp recording from a GFP-SynGAP-expressing neuron. This recording was obtained from a neuron adjacent to that in Cc1. (Calibration: 250 ms, 100 pA.) (D) Normalized AMPAR-mEPSCs from GFP-SynGAP- or eGFP-expressing neurons. To arrive at the normalized values, averages of frequency and amplitudes of mini events were taken from each recorded neuron. The average frequency (Hz) and amplitude (pA) from each neuron were then averaged to give a value for the entire population (untransfected = 2.59 ± 0.46 Hz, 18.8 ± 0.91 pA, n = 11; GFP-SynGAP = 0.37 ± 0.09 Hz, 9.77 ± 0.91 pA, n = 11). The average of the transfected population was normalized to the average of the untransfected (control) population to illustrate the overall effect of the expressed protein on each mEPSC parameter. Hence, values less than one represent a decrease in overall synaptic function, whereas values greater than one represent an increase. Statistical significance was determined by a Student's t test (two-tailed). n corresponds to the number of neurons in each population. This methodology is applied to all subsequent mEPSC plots. ∗, P < 0.001.