Abstract
Homologous recombination (HR) and translesion synthesis (TLS) are two pathways involved in the tolerance of lesions that block the replicative DNA polymerase. However, whereas TLS is frequently error-prone and, therefore, can be deleterious, HR is generally error-free. Furthermore, because the recombination enzymes and alternative DNA polymerases that perform TLS may use the same substrate, their coordination might be important to assure cell fitness and survival. This study aimed to determine whether and how these pathways are coordinated in Escherichia coli cells by using conjugational replication and recombination as a model system. The role of the three alternative DNA polymerases that are regulated by the SOS system was tested in DNA polymerase III holoenzyme-proficient and -deficient mutants. When PolIII is inactive, the alternative DNA polymerases copy DNA in the following order: PolII, PolIV, and PolV. The observed hierarchy corresponds to the selective constraints imposed on the genes coding for alternative DNA polymerases observed in natural populations of E. coli, suggesting that this hierarchy depends on the frequency of specific damages encountered during the evolutionary history of E. coli. We also found that DNA replication and HR are in competition and that they can precede each other. Our results suggest that there is probably not an active choice of which pathway to use, but, rather, the nature and concentration of lesions that lead to formation of ssDNA and the level of SOS induction that they engender might determine the outcome of the competition between HR and alternative DNA polymerases.
Keywords: SOS, translesion synthesis, polB, dinB, umuDC
All living organisms possess inducible genetic networks capable of responding to, and coping with, external and endogenous stresses. The paradigm for such a network is the Escherichia coli SOS system (1) that is induced in response to stresses that damage DNA and/or interfere with the replication catalyzed by DNA polymerase III holoenzyme (PolIII) (2). All these stresses increase the intracellular concentration of ssDNA, which is the SOS-inducing signal (2). ssDNA is the substrate for RecA protein to which it binds and forms a RecA-nucleofilament (RecA*). Depending on the nature of the DNA substrate, loading of RecA requires either RecFOR or RecBCD complexes. The RecFOR complex facilitates the formation of RecA* on ssDNA gaps covered with ssDNA-binding protein (3), whereas the substrate for the RecBCD complex is a blunt, or nearly blunt, dsDNA end from which it produces ssDNA onto which it loads RecA (4). RecA* acts as a coprotease, promoting the self-cleavage of the SOS repressor LexA, thus inducing the SOS response (5). At least 40 genes belong to the SOS regulon, some of which are involved in pathways for DNA repair and tolerance of lesions, like homologous recombination (HR) and translesion synthesis (TLS) (6, 7).
E. coli possesses three alternative DNA polymerases, regulated by the SOS system capable of performing TLS: PolII, PolIV, and PolV (encoded by polB, dinB, and umuDC genes, respectively) (8–12). When a lesion blocks the PolIII, alternative DNA polymerases can replace it and copy the DNA template across the lesion, after which PolIII can resume the processive replication of the chromosome (13). However, when the alternative DNA polymerases interfere with the processive chromosomal replication, they can be mutagenic or even toxic (14, 15); therefore, their activities have to be tightly regulated in the cell. Moreover, because the same lesion can be the substrate for several alternative DNA polymerases, the mechanism by which they are coordinated at the lesion site to determine which alternative DNA polymerase will act is also likely to be subject to regulation (16, 17).
The first known factor for the coordination of alternative DNA polymerases is their intracellular concentration, as suggested by the observation that the relative concentrations of PolII and PolV determine which polymerase will catalyze TLS at certain lesions (18). The intracellular concentration of each SOS DNA polymerase changes during SOS induction. In the uninduced cell, there are ≈50 molecules of PolII and 250 of PolIV, whereas PolV is undetectable (19–21). After SOS induction, polB and dinB are among the first genes induced, and the number of PolII and PolIV rapidly increases to 250 and 2,500 molecules per cell, respectively (9, 19, 20). PolV is functionally up-regulated more slowly, reaching ≈60 molecules per cell 1 hour after SOS induction (22).
The second factor important for recruitment and coordination of the alternative DNA polymerases is their binding to the β-clamp, which is required for TLS (23) and to increase the processivity of all E. coli DNA polymerases (24–28). All five E. coli DNA polymerases bind and compete for the same binding domain of the β-clamp, suggesting that this binding may mediate the coordination among DNA polymerases (29, 30).
In E. coli, a blocked replication fork can be the substrate for not only alternative DNA polymerases but also for HR enzymes that can allow replication to resume by eliminating the lesion from the template DNA strand (31, 32). Although TLS and HR can use the same substrate, their activities do not have the same impact on genome stability. Whereas TLS is error-prone and, therefore, can be deleterious, HR is generally error-free. The coordination of these two pathways might be required for an efficient recovery of the cell from DNA damages and the tuning of mutation rates. The complexity of the interplay between HR and TLS is exemplified by the fact that SOS induction, HR, and PolV-catalyzed TLS, although mutually exclusive (33–37), all depend on RecA* (4, 28, 38).
To determine the interplay among DNA polymerases and between replication and HR in vivo, we used the bacterial conjugation as a model system. During conjugation, DNA is transferred from an Hfr donor into a recipient cell as ssDNA with a leading 5′ end; it is immediately copied and can recombine with the recipient chromosome. We measured the kinetics of the appearance of the replicated DNA strand and the kinetics of its recombination and observed that there is a hierarchy of the alternative DNA polymerases in the absence of the catalytic subunit of PolIII (DnaE encoded by the dnaE gene) and that recombination and replication are in competition in the cell.
Results
Replication and Recombination in PolIII-Proficient Strain.
We studied the effects of E. coli DNA polymerases on replication and recombination of DNA transferred by conjugation in vivo and in real time (39) by using assays that measure the amount of transcribable lacZ gene in the recipient strain, even in nonviable mutants (e.g., dnaEts strain at 42°C).
For replication assays, we measured the zygotic induction of lacZ in the recipient. The donor strain contained a lacIS allele, coding for a LacIs hyperrepressor (40) and a functional lacZ gene. In this strain, almost no β-galactosidase is produced, even in the presence of isopropyl β-d-thiogalactoside (control curves on figures). During conjugation, donor DNA is transferred as a single strand to the lacZ− recipient cell and converted to dsDNA. When the donor lacZ promoter is double-stranded, the gene can be transcribed and β-galactosidase measured for about 1 h, the time needed for LacIs to accumulate in recipient cells and repress lacZ (39).
For the recombination assay, the Hfr donor lacZ gene contains a C-terminal deletion, whereas the recipient lacZ gene carries an N-terminal deletion (39). Between these two nonoverlapping deletions, there is 2.7-kb-long homology. After transfer of donor DNA in the recipient, one event of recombination between these two deletions restores a functional lacZ gene. If the lacZ promoter is replicated, the gene can be transcribed and β-galactosidase produced. Therefore, this assay allows detection of transcribable recombination products. For these crosses, the background signal was measured by using a donor strain carrying the same deletion as recipient strains (control curves on figures).
The replication signal in the recA mutant is low at 37°C and 42°C, whereas inactivation of recD in this mutant increases the replication signal at both temperatures (see Fig. 6, which is published as supporting information on the PNAS web site) (39). In recF, recB, and recB recF strains, replication is slightly or not affected at 37°C and 42°C, respectively (Fig. 6) (39). At both temperatures, recombination is moderately and strongly decreased in recF and recB mutants, respectively, relative to WT, whereas no recombination is observed for the recA, recA recD, and recB recF mutants (Fig. 6) (39, 41, 42). The β-galactosidase produced by recombination does not interfere with the measurements of replication efficiency, as suggested by the observation that WT (recombination-proficient) and recB recF (recombination-deficient) strains have similar replication efficiency (Fig. 6).
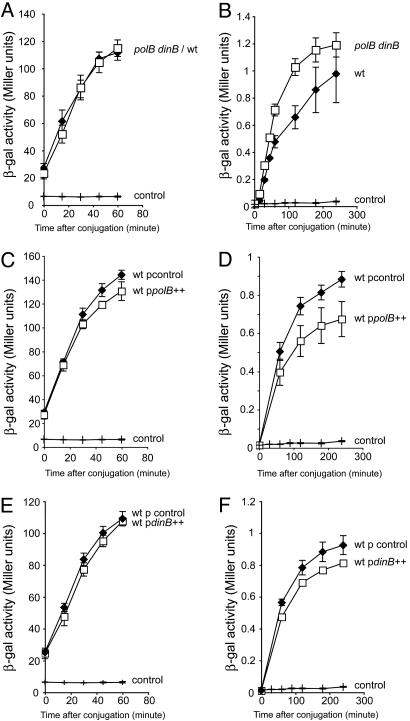
We tested the role of the alternative DNA polymerases in replication and recombination of the incoming DNA by inactivating the polB, dinB, and umuDC genes in all combinations in the PolIII-proficient recipient strain. The replication is not affected in any of the seven combinations of single, double, or triple mutants (Fig. 1A for the polB dinB double mutant and data not shown for the other mutants). However, a small but significant increase in the amount of transcribable recombination products is observed in polB dinB and polB dinB umuDC mutants relative to the WT (Fig. 1B and data not shown). Conversely, overproduction of PolII and, to a lesser extent, PolIV, from low-copy plasmids decreases the production of transcribable recombination products without affecting replication (Fig. 1 C–F). This effect is not observed in strains overproducing PolII and PolIV that contain mutations in the β-clamp-binding domain (data not shown).
Fig. 1.
Role of PolII and PolIV in DnaE-proficient strain. (A, C, and E) The kinetics of replication. (B, D, and F) The kinetics of the appearance of transcribable recombination products at 37°C. For replication and recombination assays, donor strains are Hfr3000 proC::Tn5lacIS and Hfr3000 lacZΔT(CmR), respectively. Recipients are derived from MG1655 nalR lacZΔP, and their genotypes are indicated next to their respective curves. Each point represents the mean (±SE) values from at least three independent experiments. One Miller’s unit is the amount of β-galactosidase that produces 1 nmol of orthonitrophenol per minute at 28°C, pH 7.0 (51).
Replication and Recombination in dnaEts Strain.
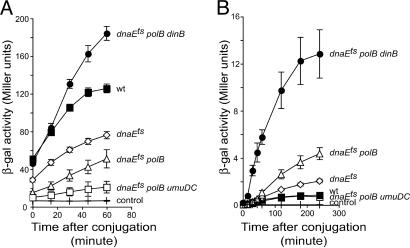
The replication of transferred DNA in a dnaEts recipient strain at 42°C is decreased, but not abolished, relative to the PolIII-proficient strain (Fig. 2A). The decrease in replication in the dnaEts recipient strain leads to an increase in the amount of transcribable recombination products (Fig. 2B). The β-galactosidase produced by recombination in the dnaEts mutant does not interfere with the measurements of replication efficiency, as suggested by the observation that inactivation of the recF or recB gene product decreases the amount of transcribable recombination products, whereas replication is slightly or not affected, respectively (see Fig. 7 A and B, which is published as supporting information on the PNAS web site). The remaining replication observed in the dnaEts mutant at 42°C is due to the activity of alternative DNA polymerases. Although the absence of PolIV and/or PolV does not affect the replication or recombination in dnaEts background (data not shown), replication is reduced, and the amount of transcribable recombination products is increased in the absence of PolII (Fig. 2 A and B). In a dnaEts polB strain, whereas inactivation of umuDC reduces the replication efficiency of the incoming DNA, inactivation of dinB increases it above the level observed for the WT strain (Fig. 2A). The replication in a dnaEts polB dinB strain does not depend on PolV, because replication in dnaEts polB dinB umuDC and dnaEts polB dinB strains is similar (data not shown). These results suggest that the alternative DNA polymerases participate in the conjugational DNA replication in dnaEts background at 42°C.
Fig. 2.
Replication and recombination in DnaE-defective strains. (A) The kinetics of replication. (B) The kinetics of the appearance of transcribable recombination products at 42°C. For replication and recombination assays, donor strains are Hfr3000 proC::Tn5 lacIS and Hfr3000 lacZΔT(CmR), respectively. Recipients are derived from MG1655 nalR lacZΔP, and their genotypes are indicated next to their respective curves. Each point represents the mean (±SE) values from at least three independent experiments.
Alternative DNA Polymerase and Chromosomal DNA Replication.
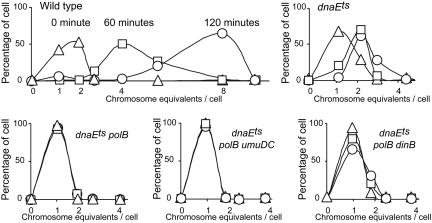
To verify the relevance of our results for chromosomal replication, we analyzed, by flow cytometry, the DNA content in the WT and different DNA polymerase mutant cells during incubation at 42°C with an inhibitor of cell division. After 2 hours of incubation, the WT strain reaches eight chromosome equivalents per cell (Fig. 3). In a dnaEts mutant, replication is reduced, and PolII performs the remaining replication (Fig. 3). No difference is observed between the dnaEts polB and dnaEts polB umuDC mutants. In the dnaEts polB dinB mutant, the percentage of cells with two chromosome equivalents increases during incubation, suggesting that some DNA replication is ongoing (Fig. 3). These results show that the alternative DNA polymerases can participate in chromosomal DNA replication in dnaEts background at 42°C.
Fig. 3.
Chromosomal replication in the WT and different DNA polymerases mutants. Flow-cytometric analyses of the DNA content of the MG1655 nalR lacZΔP and its dnaEts, dnaEts polB, dnaEts polB umuDC, and dnaEts polB dinB derivatives after incubation at 42°C for 0 (▵), 1 (□), and 2 h (○) with an inhibitor of cell division (cephalexin). This figure represents the results of one of three experiments that all showed the identical pattern of the effects of different polymerases on chromosomal DNA content.
Role of the β-Clamp in DNA Replication and Recombination.
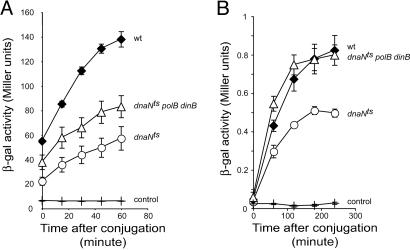
We tested the role of the β-clamp using a dnaNts allele at restrictive temperature and observed that replication efficiency and the amount of transcribable recombination products in this mutant are decreased relative to the WT strain (Fig. 4). The reduction in replication efficiency is consistent with the suggestion that this mutant protein is impaired in its interaction with DnaE (43). Inactivation of both polB and dinB genes in the dnaNts strain showed an increase in replication efficiency, suggesting that, in this genetic background, PolII and PolIV compete with (an)other DNA polymerase(s) to perform DNA replication (Fig. 4).
Fig. 4.
Replication and recombination in a β-clamp-defective strain. (A) The kinetics of replication. (B) The kinetics of the appearance of transcribable recombination products at 42°C. For replication and recombination assays, donor strains are Hfr3000 proC::Tn5 lacIS and Hfr3000 lacZΔT(CmR), respectively. Recipients are derived from MG1655 nalR lacZΔP, and their genotypes are indicated next to their respective curves. Each point represents the mean (±SE) of values from at least three independent experiments.
Interplay Between Replication and Recombination.
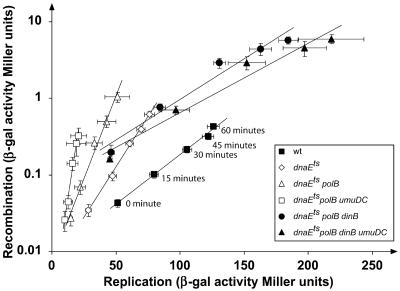
Our recombination assay can detect the recombination event between two lacZ alleles only if donor lacZ promoter has been replicated. Therefore, the absolute value of detected transcribable recombination products depends on the absolute value of replication, not only because replication provides more or less substrate for recombination, but also by allowing detection of recombination events. For example, the dnaEts polB umuDC strain showed the lowest replication efficiency and amount of transcribable recombination products (Fig. 2 A and B) but has the strongest hyperrecombination phenotype per unit of replicated DNA of all examined strains (Fig. 5; and see Table 1, which is published as supporting information on the PNAS web site). The same is true for dnaNts strain at restrictive temperature, which presents a hyperrecombination phenotype per unit of replicated DNA (see Fig. 8, which is published as supporting information on the PNAS web site, and Table 1).
Fig. 5.
Relationship between replication and recombination. The data used for this analysis are measurements of replication and recombination efficiencies during the 1st hour after mating interruption. Each point represents the mean (±SE) of values from at least four independent experiments. Slope of the correlation between the replication and the recombination and coefficient of determination (R2) are, for WT, 0.029 and 0.99; for dnaEts, 0.061 and 0.99; for dnaEts polB, 0.1 and 0.99; for dnaEts polB umuDC, 0.25 and 0.98; for dnaEts polB dinB, 0.024 and 0.96; and, for dnaEts polB dinB umuDC, 0.021 and 0.96.
Consequently, instead of comparing absolute values of transcribable recombination products, we correlated the rate of formation of transcribable recombination products and the rate of DNA replication. For all strains, we observed that the recombination rate increases exponentially with the replication rate (Fig. 5). However, the slopes of these correlations observed for different strains are very different (Figs. 5 and 8, and Table 1). Relative to the WT strain, mutants that show a decrease in the rate of replication (dnaEts, dnaEts polB, and dnaEts polB umuDC strains) generate more recombinants per amount of replicated DNA per unit of time. In contrast, strains that show an increase in the rate of replication relative to the WT strain (dnaEts polB dinB and dnaEts polB dinB umuDC strains) generate less recombinant per amount of replicated DNA per unit of time (Fig. 5). These results suggest that the ssDNA ahead of the replication site is subject to a constant competition among DNA polymerases, and between these DNA polymerases and the recombination enzymes.
Discussion
This study aimed to determine the interplay among alternative DNA polymerases and between replication and HR in E. coli. To address these questions, we measured the replication and recombination of the DNA transferred by conjugation in vivo.
Hierarchy of Alternative DNA Polymerases in Conjugational Replication.
We observed that there is a hierarchy among different DNA polymerases for conjugative DNA replication. The first polymerase is PolIII, which, when inactivated, is replaced by PolII (Fig. 2A). In the dnaEts polB strain, inactivation of umuDC reduces replication, whereas the inactivation of dinB increases replication independently whether PolV is present or not (Fig. 2A). These data may be explained, as previously proposed for TLS across bulky lesions (16, 17), by the cooperation between PolIV and PolV for replicating DNA. In our experiments, cooperation between PolIV and PolV in dnaEts polB background may take several nonexclusive forms. In the absence of PolIII and PolII, PolIV may have access to the β-clamp and synthesize a short stretch of DNA that could be elongated only and specifically by PolV. Another possibility is that PolV or its precursors assist PolIV-catalyzed DNA synthesis. Alternatively, PolIV could prevent access to the replication site to DNA polymerases other than PolV to perform synthesis. In the dnaEts polB strain, according to all these hypotheses, PolIV has first access to replication site and prevents (an)other DNA polymerase(s) from performing the DNA synthesis observed in dnaEts polB dinB and dnaEts polB dinB umuDC mutants, whereas inactivation of umuDC would reduce the capacity of the cell to catalyze DNA synthesis. So the hierarchy for conjugational replication might be PolIII, PolII, PolIV, and PolV. Similarly, the study of the mutagenesis in a dnaEts strain led to the conclusion that PolII is the first polymerase that has access to a replication fork when DNA polymerase PolIII is defective, whereas the following one is PolIV (44). The observed hierarchy does not reflect the relative number of each DNA polymerase in the SOS-noninduced or -induced cells. Therefore, it may be that the affinity of binding domains of different alternative DNA polymerases for the β-clamp plays a determining role in establishing which polymerase would be used.
PolII and PolIV Might Be the Major Competitors of DnaE.
Which DNA polymerase replicates the DNA in dnaEts polB dinB and dnaEts polB dinB umuDC mutants? One candidate is PolI, the only DNA polymerase still functional in this strain. Indeed, it is reported that PolI can replicate DNA with high processivity (45). However, we were unable to test this hypothesis because it was impossible to construct a dnaEts polB dinB polA mutant at permissive temperatures. Another candidate is DnaEts itself. It has been proposed that the DnaE486 protein has a defect in its ability to bind to the β-clamp (46). Therefore, at the restrictive temperature, the DnaE486 could lose the competition against PolII and PolIV for binding to the β-clamp. Once the polB and dinB genes are inactivated, the DnaE486 could bind again to the β-clamp and, hence, participate in the replication of DNA.
The hypothesis that PolII and PolIV outcompete PolIII whenever its interaction with β-clamp is perturbed is corroborated by our observation that the inactivation of both polB and dinB genes in a dnaNts (β-clamp-impaired in its interaction with DnaE) (43) strain at restrictive temperature results in an increase of replication (Fig. 4). Similarly, PolII and PolIV are responsible for the toxicity in a holDts strain at restrictive temperature (holD codes for a component of the DNA polymerase III holoenzyme) (15). Therefore, our results confirm that these two DNA polymerases interfere with DNA replication when interactions between different PolIII subunits are not optimal.
Involvement of PolV in Conjugational Replication.
How can we reconcile our observation that PolV does not significantly participate in DNA synthesis when PolIII, PolII, and PolIV are not active (in strongly SOS-induced cells) with reports that PolV is a very efficient DNA polymerase capable of bypassing a wide spectrum of different replication-blocking lesions in vivo and in vitro (14)? PolV is the only E. coli alternative DNA polymerase whose activity is transcriptionally and posttranslationally controlled to assure its presence only in cells that have suffered DNA damage that cannot be processed by other DNA repair systems (47). Therefore, it may be that, in vivo, yet another level of control of PolV activity exists that prevents the performing of error-prone DNA synthesis in the absence of specific types of DNA lesions, as is the case in our study. Alternatively, in vivo PolV processivity may be so low in this strain that the amount of synthesized DNA is below the detection threshold of our assay. Finally, we cannot exclude that the temperature shift required for inactivation of thermosensitive mutants induces a “heat-shock” response that also modifies PolV stability and activity.
Involvement of Alternative Polymerases in Chromosomal Replication.
Is the observed interplay between alternative polymerases specific to conjugational replication? Despite the fact that, during conjugation, just one strand is transferred, its replication depends on DnaE and DnaN but also on the replicative helicase DnaB, suggesting that most of the components (if not all) involved in the replication of the chromosome might be required for this replication (Figs. 2A, 4, and 6C) (48). Moreover, we observed that the hierarchy and the interplay among DNA polymerases are partially valid for the chromosomal replication (Fig. 3), i.e., PolIII is replaced by PolII, whereas inactivation of the dinB gene in the dnaEts polB strain allows DNA replication to restart. Only the effect of umuDC inactivation was not detected. However, if the effects of different alternative DNA polymerases on conjugational and chromosomal replication seem to be similar, suggesting that the same hierarchy might apply on the chromosome, they are quantitatively different. These differences might be due to problems of coordination and/or stability of replisome subunits during replication of both chromosomal DNA strands compared with conjugational replication, i.e., replication of only one strand.
Hierarchy of Selective Constraints on Alternative Polymerases.
The existence of a hierarchy among different TLS polymerases is also suggested by the analysis of the patterns of DNA sequence diversity for polB, dinB, and umuC genes from E. coli natural isolates. Because natural selection fine-tunes the rate of evolution of genes according to their functions, the high conservation of genes coding for alternative DNA polymerases suggests that their activity is very important for cell fitness and survival (49). The ratio of the number of nonsynonymous substitutions (KA) and of the number of synonymous substitutions (KS), giving an indication of relative intensity of the selective constraint on replacement substitutions, showed that polB (KA/KS = 0.0041) is more constrained than dinB (KA/KS = 0.019), whereas umuC (KA/KS = 0.19) appears to be subject to the lowest constraint. Because alternative DNA polymerases participate in the processing of DNA lesions (14), the level of selective constraint may reflect the frequency by which they are required to assure survival. It is striking to note that the constraints imposed on the alternative polymerases during E. coli evolution correspond to the hierarchy observed in our experiments in the absence of exogenously induced lesions. Therefore, the recruitment of alternative polymerases might follow a hierarchy that reflects the frequency of specific damages encountered during the evolutionary history of E. coli.
Interplay Between Replication and Recombination.
Because the recombination enzymes and different alternative DNA polymerases may use the same substrate to ensure the tolerance of lesions that block the progression of the DNA PolIII, their coordination might be important to assure cell fitness and survival (14, 31, 32). Indeed, the result of their activity can be very different, because HR is, in general, error-free, whereas TLS is generally error-prone. We found that, in E. coli, the recombination rate of DNA transferred by conjugation is a function of its replication rate, i.e., the decrease/increase of replication rate results in the increase/decrease of recombination rate (Fig. 5). This result suggests that the ssDNA ahead of the replication site is constantly subject to a competition between replication and recombination enzymes. This competition is also illustrated by the observation that the inactivation of polB and dinB increases the efficiency of recombination, whereas their overexpression decreases it (Fig. 1), suggesting that some difficulties encountered during DNA replication are easily bypassed by an alternative DNA polymerase (such as PolII and/or PolIV), hence, proceeding and precluding HR, whereas, in their absence, HR might act. There might not be an active choice of which pathway to use, but, rather, the nature and concentration of lesions that lead to an increase in the number, the length and the lifetime of ssDNA gaps, and the level of SOS induction that they engender could determine the outcome of the competition between HR and alternative DNA polymerases.
Materials and Methods
Donor Strains.
All donor strains were Hfr3000 derivatives (50). For recombination experiments, donor strains were Hfr3000 lacZΔT(CmR) and Hfr3000 lacZΔP(CmR). The construction of the lacZΔT(CmR), lacZΔP(CmR), and lacZΔP alleles is described in ref. 39. For replication experiments, the donor strain was Hfr3000 proC::Tn5 lacI694 (designated as lacIS in the text) (39).
Recipient Strains.
All recipient strains were MG1655 derivatives (50). We selected a spontaneous nalidixic acid-resistant mutant, introduced the lacZΔP(CmR) allele, and removed the CmR cassette. This MG1655 nalR lacZΔP strain was designated as WT. All other alleles and combination of alleles were introduced in this strain by P1-mediated transduction (51). The alleles used were: recA938::Tn9–200 (52), recD1901::Tn10 (CGSC: 7429), recB268::Tn10 (53), ΔrecB::PhleoR (constructed by I. Bjedov, Institut National de la Santé et de la Recherche Médicale U571), recF400::Tn5 (provided by B. Michel, Centre de Génétique Moléculaire, Gyf-sur-Yvette, France), dnaE486 zae502::Tn10 (54) (designated as dnaEts in the text), dnaN159 zid::Tn10 (55) (designated as dnaNts in the text), dnaB8 mal::Tn9 (56), dinB::KanR (57), polBΩ (Sm-Sp) (58), and ΔumuDC595::CmR (59). In strains carrying the dnaE486 zae502::Tn10 allele, the ΔrecB::PhleoR allele, which is a complete deletion of the ORF, was introduced. No differences in kinetics of replication and recombination were observed between the recB268::Tn10 and ΔrecB::PhleoR alleles introduced in the WT strain. The plasmids overexpressing PolII and PolIV, with or without capacity to bind to the β-clamp, and their respective plasmid controls are described in ref. 23.
Phenotypes were confirmed by antibiotic resistance and, when appropriate, UV sensitivity, thermosensitivity, or PCR. Antibiotics were used at the following concentrations: kanamycin (100 μg/ml), spectinomycin (75 μg/ml), tetracycline (12.5 μg/ml), phleomycin (10 μg/ml), chloramphenicol (30 μg/ml), cephalexin (12 μg/ml), rifampicin (100 μg/ml), and nalidixic acid (40 μg/ml).
Conjugational Crosses and β-Galactosidase Assays.
Overnight cultures were diluted 50-fold, in LB for donor cells, or LB supplemented with isopropyl β-d-thiogalactoside (IPTG) (100 μM) for recipient cells, and grown to ≈108 cells per ml. Two milliliters of donor and 2 ml of recipient culture were then mixed and deposited on nitrocellulose filter (0.45-μm pore size; Schleicher & Schuel). For kinetics at 42°C, recipients were incubated with shaking at 42°C for 15 min before conjugation. Filters were deposited on a prewarmed LB plate containing 100 μM IPTG for 40 min at 37°C or 42°C. After this period, corresponding to the time 0 of the experiment, cells were resuspended by vortexing for 1 minute in 2 ml of M9 minimal medium supplemented with thiamine (30 μg/ml), uracil (0.002%), MgSO4 (3 mM), casaminoacids (0.1%) (as a poor carbon source to reduce cell growth to a minimum), IPTG (100 μM), and nalidixic acid to stop the conjugation and kill donor cells. Cell suspensions were maintained at 37°C or 42°C with agitation, and samples were withdrawn at appropriate time points. β-galactosidase activity was measured as described in ref. 51. To measure low levels of β-galactosidase activities, longer incubation times were necessary (up to 24 h), always accompanied with the control for spontaneous hydrolysis of O-nitrophenyl-β-d-galactoside. When recipient strains carried a plasmid, the appropriate antibiotic was added in all liquid medium steps but not in the conjugation plate.
Flow Cytometry.
Overnight cultures were diluted 500-fold in LB and grown at 30°C until they reached an OD600 of 0.2. At this time point, corresponding to the time 0 of the experiment, cephalexin, an inhibitor of cell division, was added to cultures. These cultures were incubated at 42°C, and samples were taken at time 0, 1, and 2 h, resuspended in 10 mM Tris·Mg2+, pH 7.4, fixed in 80% methanol, and stained with DAPI (0.3 μg/ml) and 20,000 cells were analyzed by flow cytometry. The numbers of chromosomal equivalents per cell for different strains were estimated relative to the number of chromosomal equivalents per cell of the WT strain treated with cephalexin and rifampicin.
Estimations of the Number of Synonymous and Nonsynonymous Substitutions.
Partial sequences of polB (977-bp), dinB (1,010-bp), and umuC (1,230-bp) genes from a set of 30 strains representative of the genetic diversity of the E. coli reference (ECOR) collection of natural isolates were retrieved from GenBank (AF483912–AF483939 for polB, AF483080–AF483106 for dinB, and AF483970–AF483998 for umuC) (49). The estimations of the number of nonsynonymous substitutions per nonsynonymous site (KA) and the number of synonymous substitutions per synonymous site (KS) have been performed by using the program dnasp (60), as described in ref. 61.
Supplementary Material
Acknowledgments
We thank M. Radman for helpful comments on the manuscript; M. Selva for technical assistance; M. C. Gendron (Institut Jacques Monod, Paris) for flow-cytometry analysis; and B. Michel, O. Becherel [Centre Nationale de la Recherche Scientifique (CNRS), UPR9003, Strasbourg, France], and J. Wagner (CNRS) for the generous gift of strains and plasmids. S.D. is a “Fondation pour la Recherche Médicale”, Ministère de l’Éducation Nationale, de la Recherche et de la Technologie and a “Ligue Contre le Cancer” Fellow.
Abbreviations
- HR
homologous recombination
- PolIII
DNA polymerase III holoenzyme
- TLS
translesion synthesis.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sutton M. D., Smith B. T., Godoy V. G., Walker G. C. Annu. Rev. Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- 2.Sassanfar M., Roberts J. W. J. Mol. Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 3.Morimatsu K., Kowalczykowski S. C. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 4.Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little J. W., Edmiston S. H., Pacelli Z., Mount D. W. Proc. Natl. Acad. Sci. USA. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courcelle J., Khodursky A., Peter B., Brown P. O., Hanawalt P. C. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez De Henestrosa A. R., Ogi T., Aoyagi S., Chafin D., Hayes J. J., Ohmori H., Woodgate R. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 8.Bonner C. A., Hays S., McEntee K., Goodman M. F. Proc. Natl. Acad. Sci. USA. 1990;87:7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonner C. A., Randall S. K., Rayssiguier C., Radman M., Eritja R., Kaplan B. E., McEntee K., Goodman M. F. J. Biol. Chem. 1988;263:18946–18952. [PubMed] [Google Scholar]
- 10.Reuven N. B., Arad G., Maor-Shoshani A., Livneh Z. J. Biol. Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 11.Tang M., Shen X., Frank E. G., O’Donnell M., Woodgate R., Goodman M. F. Proc. Natl. Acad. Sci. USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner J., Gruz P., Kim S. R., Yamada M., Matsui K., Fuchs R. P., Nohmi T. Mol. Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg E. C., Wagner R., Radman M. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 14.Goodman M. F. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 15.Viguera E., Petranovic M., Zahradka D., Germain K., Ehrlich D. S., Michel B. Mol. Microbiol. 2003;50:193–204. doi: 10.1046/j.1365-2958.2003.03658.x. [DOI] [PubMed] [Google Scholar]
- 16.Napolitano R., Janel-Bintz R., Wagner J., Fuchs R. P. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner J., Etienne H., Janel-Bintz R., Fuchs R. P. DNA Repair (Amsterdam) 2002;1:159–167. doi: 10.1016/s1568-7864(01)00012-x. [DOI] [PubMed] [Google Scholar]
- 18.Becherel O. J., Fuchs R. P. Proc. Natl. Acad. Sci. USA. 2001;98:8566–8571. doi: 10.1073/pnas.141113398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S. R., Matsui K., Yamada M., Gruz P., Nohmi T. Mol. Genet. Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- 20.Qiu Z., Goodman M. F. J. Biol. Chem. 1997;272:8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- 21.Woodgate R., Ennis D. G. Mol. Gen. Genet. 1991;229:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- 22.Sommer S., Boudsocq F., Devoret R., Bailone A. Mol. Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 23.Becherel O. J., Fuchs R. P., Wagner J. DNA Repair (Amsterdam) 2002;1:703–708. doi: 10.1016/s1568-7864(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 24.Lopez de Saro F. J., O’Donnell M. Proc. Natl. Acad. Sci. USA. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruck I., O’Donnell M. Genome Biol. 2001;2:REVIEWS3001. doi: 10.1186/gb-2001-2-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonner C. A., Stukenberg P. T., Rajagopalan M., Eritja R., O’Donnell M., McEntee K., Echols H., Goodman M. F. J. Biol. Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 27.Wagner J., Fujii S., Gruz P., Nohmi T., Fuchs R. P. EMBO Rep. 2000;1:484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham P., Bertram J. G., O’Donnell M., Woodgate R., Goodman M. F. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 29.Lopez de Saro F. J., Georgescu R. E., Goodman M. F., O’Donnell M. EMBO J. 2003;22:6408–6418. doi: 10.1093/emboj/cdg603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnouf D. Y., Olieric V., Wagner J., Fujii S., Reinbolt J., Fuchs R. P., Dumas P. J. Mol. Biol. 2004;335:1187–1197. doi: 10.1016/j.jmb.2003.11.049. [DOI] [PubMed] [Google Scholar]
- 31.Friedberg E. C., Walker G. C., Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 32.Michel B., Grompone G., Flores M. J., Bidnenko V. Proc. Natl. Acad. Sci. USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boudsocq F., Campbell M., Devoret R., Bailone A. J. Mol. Biol. 1997;270:201–211. doi: 10.1006/jmbi.1997.1098. [DOI] [PubMed] [Google Scholar]
- 34.Harmon F. G., Rehrauer W. M., Kowalczykowski S. C. J. Biol. Chem. 1996;271:23874–23883. [PubMed] [Google Scholar]
- 35.Rehrauer W. M., Bruck I., Woodgate R., Goodman M. F., Kowalczykowski S. C. J. Biol. Chem. 1998;273:32384–32387. doi: 10.1074/jbc.273.49.32384. [DOI] [PubMed] [Google Scholar]
- 36.Rehrauer W. M., Lavery P. E., Palmer E. L., Singh R. N., Kowalczykowski S. C. J. Biol. Chem. 1996;271:23865–23873. [PubMed] [Google Scholar]
- 37.Sommer S., Bailone A., Devoret R. Mol. Microbiol. 1993;10:963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 38.Burckhardt S. E., Woodgate R., Scheuermann R. H., Echols H. Proc. Natl. Acad. Sci. USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veaute X., Delmas S., Selva M., Jeusset J., Le Cam E., Matic I., Fabre F., Petit M. A. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willson C., Perrin D., Cohn M., Jacob F., Monod J. J. Mol. Biol. 1964;141:582–592. doi: 10.1016/s0022-2836(64)80013-9. [DOI] [PubMed] [Google Scholar]
- 41.Birge E. A., Low K. L. J. Mol. Biol. 1974;83:447–457. doi: 10.1016/0022-2836(74)90506-3. [DOI] [PubMed] [Google Scholar]
- 42.Lloyd R. G., Evans N. P., Buckman C. Mol. Gen. Genet. 1987;209:135–141. doi: 10.1007/BF00329848. [DOI] [PubMed] [Google Scholar]
- 43.Sutton M. D. J. Bacteriol. 2004;186:6738–6748. doi: 10.1128/JB.186.20.6738-6748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banach-Orlowska M., Fijalkowska I. J., Schaaper R. M., Jonczyk P. Mol. Microbiol. 2005;58:61–70. doi: 10.1111/j.1365-2958.2005.04805.x. [DOI] [PubMed] [Google Scholar]
- 45.Camps M., Loeb L. A. Cell Cycle. 2004;3:116–118. [PubMed] [Google Scholar]
- 46.Vandewiele D., Fernandez de Henestrosa A. R., Timms A. R., Bridges B. A., Woodgate R. Mutat. Res. 2002;499:85–95. doi: 10.1016/s0027-5107(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez M., Woodgate R. BioEssays. 2002;24:141–148. doi: 10.1002/bies.10040. [DOI] [PubMed] [Google Scholar]
- 48.Willets N., Wilkins B. Microbiol. Rev. 1984;48:24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjedov I., Lecointre G., Tenaillon O., Vaury C., Radman M., Taddei F., Denamur E., Matic I. DNA Repair (Amsterdam) 2003;2:417–426. doi: 10.1016/s1568-7864(02)00241-0. [DOI] [PubMed] [Google Scholar]
- 50.Bachmann B. J. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F. C., Curtiss R. III, Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., editors. Vol. 2. Washington DC: Am. Soc. Microbiol.; 1996. pp. 2460–2488. [Google Scholar]
- 51.Miller J. H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 52.Wertman K. F., Wyman A. R., Botstein D. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]
- 53.Lloyd R. G., Buckman C., Benson F. E. J. Gen. Microbiol. 1987;133:2531–2538. doi: 10.1099/00221287-133-9-2531. [DOI] [PubMed] [Google Scholar]
- 54.Grompone G., Seigneur M., Ehrlich S. D., Michel B. Mol. Microbiol. 2002;44:1331–1339. doi: 10.1046/j.1365-2958.2002.02962.x. [DOI] [PubMed] [Google Scholar]
- 55.Flores M. J., Bidnenko V., Michel B. EMBO Rep. 2004;5:983–988. doi: 10.1038/sj.embor.7400262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seigneur M., Bidnenko V., Ehrlich S. D., Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 57.Kim S.-R., Maenhaut-Michel G., Yamada M., Yamamoto Y., Matsui K., Sofuni T., Nohmi T., Ohmori H. Proc. Natl. Acad. Sci. USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Escarceller M., Hicks J., Gudmundsson G., Trump G., Touati D., Lovett S., Foster P. L., McEntee K., Goodman M. F. J. Bacteriol. 1994;176:6221–6228. doi: 10.1128/jb.176.20.6221-6228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woodgate R. Mutat. Res. 1992;281:221–225. doi: 10.1016/0165-7992(92)90012-7. [DOI] [PubMed] [Google Scholar]
- 60.Rozas J., Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 61.Nei M., Gojobori T. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.