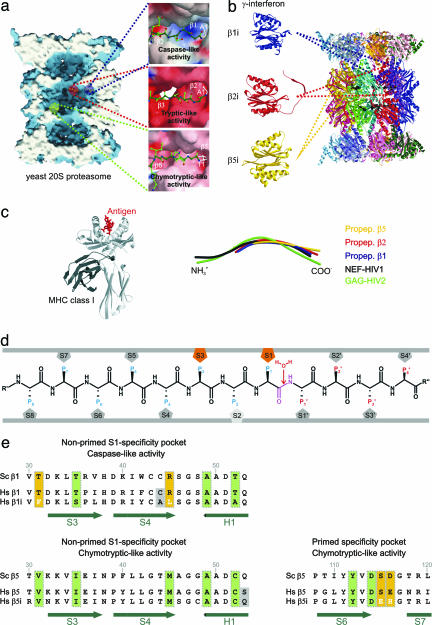
Fig. 1.
Proteasomal proteolytically active sites involved in the generation of MHC class I ligands. (a) Surface representation of the yeast 20S proteasome in complex with propeptides, clipped along the cylindrical pseudo sevenfold symmetry axis. Accessible surfaces are depicted in blue, and the cutting surface is in white. Propeptides are shown as space-filling models in yellow and indicate the proteolytically different active sites. The various proteolytic active centers are marked in a specific color coding: blue, subunit β1; red, subunit β2; and green, subunit β5. Cleavage preferences, termed caspase-, tryptic-, and chymotryptic-like activity, are zoomed and illustrated as surfaces; propeptides are presented as ball-and-stick models. Surface colors indicate positive and negative electrostatic potential contoured from 15 kT/e (intense blue) to −15 kT/e (intense red). (b) Topology of the 28 subunits of the yeast 20S proteasome in ribbon presentation. IFN-γ-inducible mammalian subunits β1i, β2I, and β5i are modeled by the corresponding constitutive yeast subunits. (c Left) MHC class I molecule in complex with an antigen (c Right). Structural superposition of propeptides β1, β2, and β5 with NEF-HIV1 and GAG-HIV2 antigen bound to MHC class I molecules. (d) Standard orientation for peptide substrates bound to the proteasomal specificity pockets. Substrates are oriented from their N to their C terminus. The scissile peptide bond is shown in magenta, flanked by the nucleophilic water molecule, which is incorporated into the product during hydrolysis. Residues on the left side of the scissile peptide bond in substrates, generating the C-terminal part in the product, are termed P sites; residues on the right side are termed P′ sites. Specificity pockets, which are responsible for ligand stabilization, are termed S and S′ pockets, respectively (11). (e) Sequence alignment of the yeast and the human constitutive subunit and immunosubunit for subunit β1 (nonprimed S1 site, Upper) and subunit β5 (nonprimed S1 and primed substrate-binding channel, Lower). Conserved residues are marked by vertical green boxes, significant variations between human constitutive subunits and immunosubunits are highlighted by yellow boxes, and variations in residues in proximity to the specificity pockets are shown against a gray background. Secondary structure elements are indicated in green.