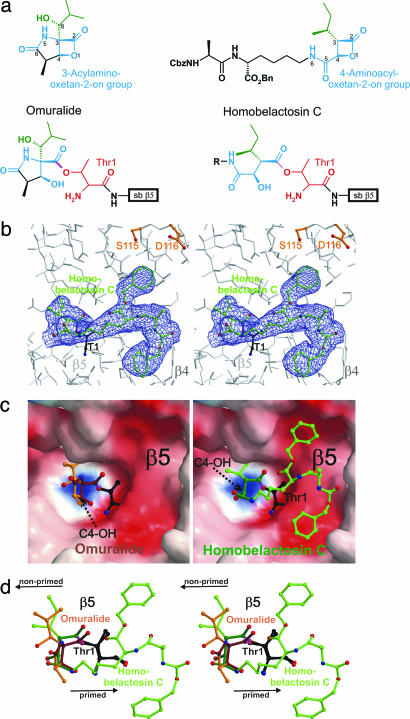
Fig. 2.
Homobelactosin C specifically binds to the chymotryptic-like active site by formation of an ester. (a) Chemical structures of omuralide and bisbenzyl-protected homobelactosin C in their native and bound conformation. The lead structure segments that in particular are involved in inhibitor binding are depicted in blue; Thr-1 of subunit β5 is in red. (b) Stereorepresentation of the chymotryptic-like active site of the yeast 20S proteasome (colored in gray) in complex with bisbenzyl-protected homobelactosin C (colored in green). Covalent linkage of the inhibitor with β5-Thr1Oγ is drawn in magenta. The electron density map (colored in blue) is contoured from 1σ around Thr-1 (colored in black) with 2Fo − Fc coefficients after twofold averaging. Temperature factor refinement indicates full occupancies of the inhibitor-binding site. The homobelactosin C derivative has been omitted for phasing. (c) Surface representation of the chymotryptic-like active site in complex with omuralide (depicted in brown, Left) and homobelactosin C (depicted in green, Right), covalently bound to Thr-1 (depicted in white). Note the overall similarity in the binding mode of both inhibitors but the different orientations of the generated C4-hydroxy group upon β-lactone ring opening (indicated by a black arrow). Surface colors indicate positive and negative electrostatic potential contoured from 15 kT/e (intense blue) to −15 kT/e (intense red). (d) Stereorepresentation of the superposition of lactacystin and bisbenzyl-protected homobelactosin C, including Thr-1 with respect to subunit β5. Omuralide is shown in brown, bisbenzyl-protected homobelactosin C is drawn in green, and the active site Thr-1 is in black. The superposition indicates that both inhibitors occupy the S1 specificity pocket in a unique way. The bisbenzyl-protected homobelactosin C is prolonged to the primed site. Nonprimed and primed sites are indicated by a black arrow.