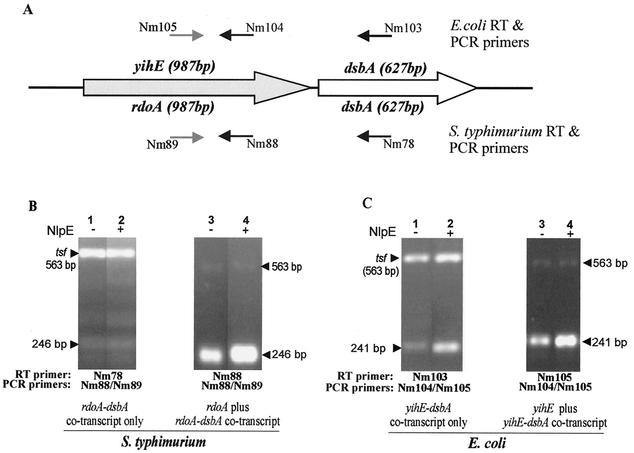
FIG. 5.
Transcript analysis of yihE, rdoA, rdoA-dsbA, and yihE-dsbA by RT-PCR. The primers utilized in the RT-PCR are illustrated in panel A, which shows the organization of the rdoA (yihE)-dsbA region in serovar Typhimurium and E. coli. Serovar Typhimurium primer Nm78 binds 400 bp downstream of the dsbA start codon, while primers Nm88 and Nm89 bind 855 and 612 bp downstream of the rdoA start codon. E. coli primer Nm103 binds 404 bp downstream of the dsbA start codon, while primers Nm105 and Nm104 bind 855 and 614 bp downstream of the yihE start codon. In panels B and C the lower amplicons are the target amplicons, while the upper bands are the tsf (encoding the elongation factor EF-Tsf) internal control amplicon used as a standard in the qualitative assessment of pND18 (NlpE)-induced (+) or pBAD18 (control vector)-induced (−) samples of serovar Typhimurium (B) and E. coli (C) RNA preparations. The size of the EF-tsf amplicon is 563 bp, while the serovar Typhimurium target amplicon for the rdoA-dsbA- and rdoA-specific transcript is 246 bp. The E. coli target amplicon for the yihE-dsbA- or yihE-specific transcript is 241 bp.