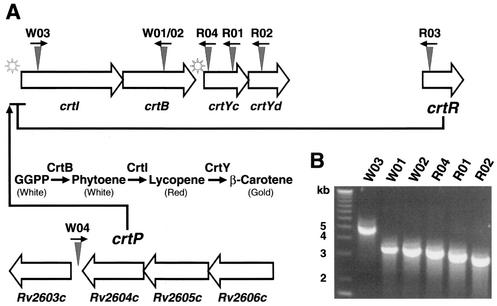
FIG. 3.
M. marinum carotenoid biosynthesis and regulatory genes, their genomic organization, and location of transposon insertions. (A) The known carotenoid biosynthesis locus in M. marinum is shown as a gene cluster from crtI to ORF-4. Below these genes the biosynthetic pathway is depicted; GGPP represents the initial substrate, geranylgeranylpyrophosphate. The novel crtR and crtP genes encode a putative repressor and a positive regulator, respectively, for the carotenoid biosynthesis locus. crtP indicates the locus containing the Rv2606c to Rv2603c homologues. The star symbols represent putative light-inducible promoters. Triangles indicate sites of transposon insertion, and arrows represent the orientation of the Kanr gene in the chromosome. R01, MmR01; R02, MmR02; R03, MmR03; R04, MmR04; W01, MmW01; W02, MmW02; W03, MmW03; W04, MmW04. (B) The location of transposon insertion in mutant MmR03 is shown relative to those in other pigmentation mutants as detected by a PCR that amplifies the genomic sequence between insertions. The distance of the transposon insertion in MmW04 from others in the carotenoid biosynthesis locus is insufficiently detected by PCR, and the results were not shown.