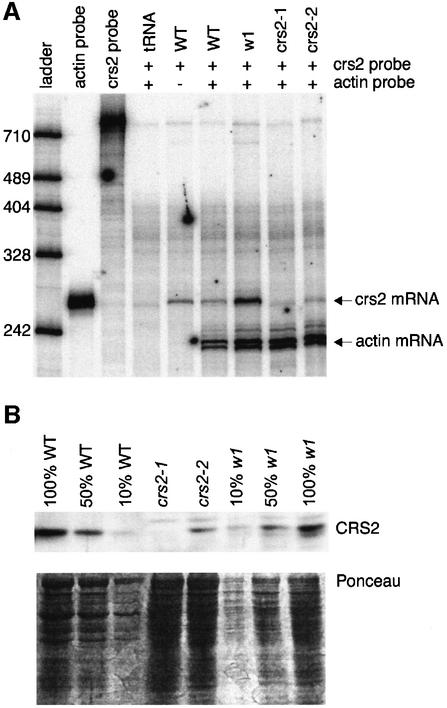
Fig. 2. Expression of the crs2 gene is disrupted in crs2-1 and crs2-2 mutants. (A) RNase-protection assay of crs2 mRNA. The probe for the crs2 mRNA was derived by in vitro transcription of the crs2 genomic sequence to the ‘right’ of the Mu1 insertion in Figure 1A. A probe complementary to a portion of the maize actin mRNA was included as an internal control. Thirty micrograms of total leaf RNA from wild-type, crs2-1, crs2-2 or w1 seedlings, or an equivalent amount of tRNA were analyzed. w1, an ivory maize mutant with a global and tight defect in chloroplast gene expression, was included to control for any effects on crs2 RNA accumulation that might be associated with defects in chloroplast development. Because the crs2 probe included intron sequence, the 253 nt fragment protected by wild-type mRNA is considerably smaller than the probe. The undigested probes (lanes 2 and 3) correspond to 1/200th as much probe as was included in each reaction. (B) Immunoblot showing CRS2 protein levels in crs2 mutants. Twenty micrograms of total leaf protein (or the indicated dilutions) from wild-type, crs2-1, crs2-2 or w1 seedlings were fractionated by SDS–PAGE. Upper panel: immunoblot probed with a CRS2 antibody. Lower panel: the same filter stained with Ponceau S to illustrate equal loading of the protein.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.