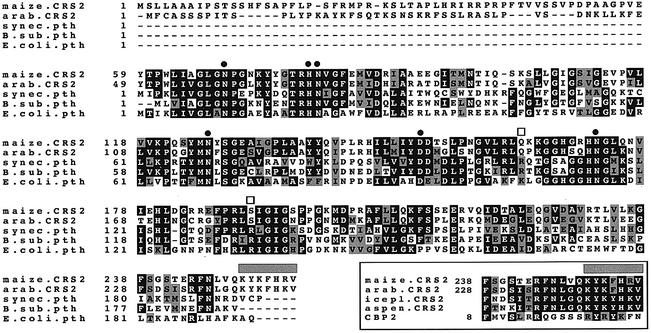
Fig. 4. Multiple sequence alignment of CRS2 with related Arabidopsis and bacterial proteins. The alignment includes sequences of the putative CRS2 ortholog in Arabidopsis (DDBJ/EMBL/GenBank accession No. BAB09443) and three bacterial PTHs (E.coli, accession No. P23932; Bacillus subtilis, accession No. P37470; Synechocystis PCC6803, accession No. Q59989). Solid circles indicate residues shown to be essential for the catalytic activity of the E.coli PTH (Schmitt et al., 1997; Fromant et al., 1999). Open squares indicate residues important for the binding of substrate by the E.coli enzyme (Fromant et al., 1999). The gray bar highlights a putative RNA-binding region that is found in predicted CRS2 proteins in plants, but not in their PTH ancestor. The lower box shows an alignment of this region of predicted plant CRS2s with a region of the yeast protein Cbp2 that has been implicated in binding group I introns and in facilitating group I intron splicing (Tirupati et al., 1999). The C-terminal sequences of the presumed CRS2 orthologs in iceplant (accession No. AI026363) and aspen (accession No. AI162403) were derived from EST sequences. Sequence alignments were calculated using ClustalW 1.8 (Thompson et al., 1994) and Boxshade (Bioinformatics group, ISREC, Lausanne, Switzerland), with default parameters.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.